5YV0
 
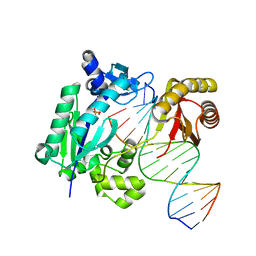 | | DNA polymerase IV - DNA ternary complex 12 | | Descriptor: | DIPHOSPHATE, DNA polymerase IV, DTN1, ... | | Authors: | Kottur, J, Nair, D.T. | | Deposit date: | 2017-11-23 | | Release date: | 2018-11-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Pyrophosphate hydrolysis is an intrinsic and critical step of the DNA synthesis reaction.
Nucleic Acids Res., 46, 2018
|
|
3HMH
 
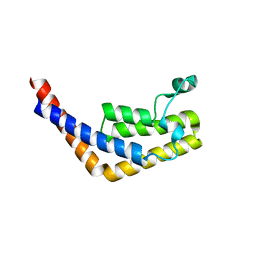 | | Crystal structure of the second bromodomain of human TBP-associated factor RNA polymerase 1-like (TAF1L) | | Descriptor: | Transcription initiation factor TFIID 210 kDa subunit | | Authors: | Filippakopoulos, P, Picaud, S, Keates, T, Zhang, Y, Pike, A.C.W, von Delft, F, Arrowsmith, C.H, Edwards, A, Weigelt, J, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-05-29 | | Release date: | 2009-06-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Histone recognition and large-scale structural analysis of the human bromodomain family.
Cell(Cambridge,Mass.), 149, 2012
|
|
5ZLV
 
 | |
3NKB
 
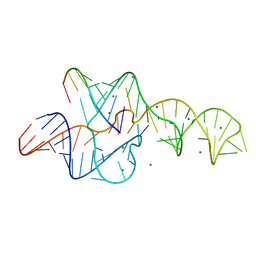 | | A 1.9A crystal structure of the HDV ribozyme precleavage suggests both Lewis acid and general acid mechanisms contribute to phosphodiester cleavage | | Descriptor: | DNA/RNA (5'-D(*(DUR))-D(*GP*G)-R(P*CP*UP*UP*GP*CP*A)-3'), MAGNESIUM ION, The hepatitis delta virus ribozyme | | Authors: | Chen, J.-H, Yajima, R, Chadalavada, D.M, Chase, E, Bevilacqua, P.C, Golden, B.L. | | Deposit date: | 2010-06-18 | | Release date: | 2010-09-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.916 Å) | | Cite: | A 1.9 A crystal structure of the HDV ribozyme precleavage suggests both Lewis acid and general acid mechanisms contribute to phosphodiester cleavage.
Biochemistry, 49, 2010
|
|
3NHG
 
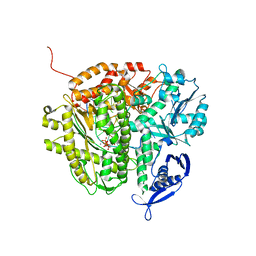 | | RB69 DNA Polymerase (S565G/Y567A) Ternary Complex with dTTP Opposite dG | | Descriptor: | CALCIUM ION, DNA (5'-D(*GP*CP*GP*GP*AP*CP*TP*GP*CP*TP*TP*AP*(DOC))-3'), DNA (5'-D(*TP*CP*AP*GP*GP*TP*AP*AP*GP*CP*AP*GP*TP*CP*CP*GP*CP*G)-3'), ... | | Authors: | Wang, M, Wang, J, Konigsberg, W.H. | | Deposit date: | 2010-06-14 | | Release date: | 2011-01-26 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Variation in Mutation Rates Caused by RB69pol Fidelity Mutants Can Be Rationalized on the Basis of Their Kinetic Behavior and Crystal Structures.
J.Mol.Biol., 406, 2011
|
|
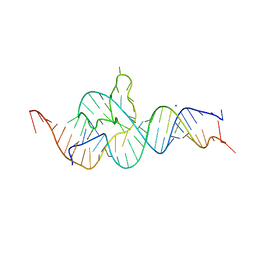
4QJD
 
 | |
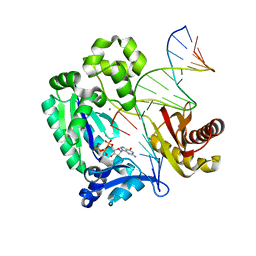
3RAX
 
 | |
8GAO
 
 | | bacteriophage T4 stalled primosome with mutant gp41-E227Q | | Descriptor: | DNA (70-mer), DNA primase, DnaB-like replicative helicase, ... | | Authors: | Feng, X, Li, H. | | Deposit date: | 2023-02-23 | | Release date: | 2023-06-07 | | Last modified: | 2023-08-23 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structural basis of the T4 bacteriophage primosome assembly and primer synthesis.
Nat Commun, 14, 2023
|
|
5OKI
 
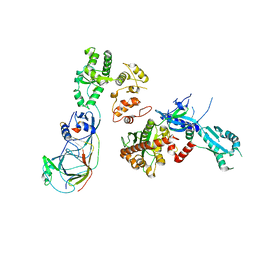 | |
8B54
 
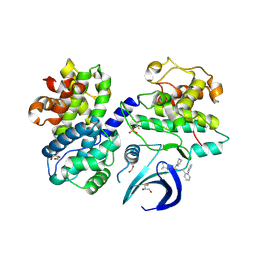 | | CDK2/cyclin A2 in complex with pyrazolo[4,3-d]pyrimidine inhibitor LGR6768 | | Descriptor: | 1,2-ETHANEDIOL, Cyclin-A2, Cyclin-dependent kinase 2, ... | | Authors: | Djukic, S, Skerlova, J, Rezacova, P. | | Deposit date: | 2022-09-21 | | Release date: | 2023-03-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Characterization of new highly selective pyrazolo[4,3-d]pyrimidine inhibitor of CDK7.
Biomed Pharmacother, 161, 2023
|
|
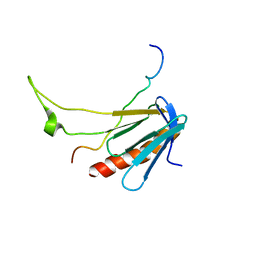
2L2I
 
 | |
5Y4Z
 
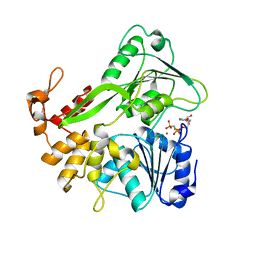 | | Crystal structure of the Zika virus NS3 helicase complex with AMPPNP | | Descriptor: | MANGANESE (II) ION, NS3 helicase, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Li, L. | | Deposit date: | 2017-08-06 | | Release date: | 2017-08-30 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural view of the helicase reveals that Zika virus uses a conserved mechanism for unwinding RNA
Acta Crystallogr.,Sect.F, 74, 2018
|
|
3NDK
 
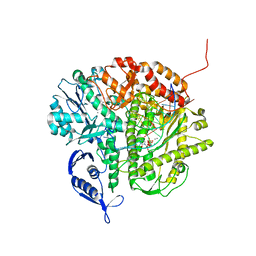 | | RB69 DNA Polymerase (Y567A) Ternary Complex with dCTP Opposite dG | | Descriptor: | 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, CALCIUM ION, DNA (5'-D(*GP*CP*GP*GP*AP*CP*TP*GP*CP*TP*TP*AP*(DOC))-3'), ... | | Authors: | Wang, M, Wang, J, Konigsberg, W.H. | | Deposit date: | 2010-06-07 | | Release date: | 2011-01-26 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Variation in Mutation Rates Caused by RB69pol Fidelity Mutants Can Be Rationalized on the Basis of Their Kinetic Behavior and Crystal Structures.
J.Mol.Biol., 406, 2011
|
|
5EFQ
 
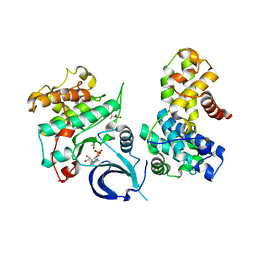 | | Crystal structure of human Cdk13/Cyclin K in complex with ADP-aluminum fluoride | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, Cyclin-K, ... | | Authors: | Hoenig, D, Greifenberg, A.K, Anand, K, Geyer, M. | | Deposit date: | 2015-10-24 | | Release date: | 2015-12-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and Functional Analysis of the Cdk13/Cyclin K Complex.
Cell Rep, 14, 2016
|
|
7VQP
 
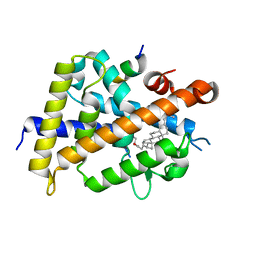 | | Vitamin D receptor complexed with a lithocholic acid derivative | | Descriptor: | 3-((R)-4-((3R,5R,8R,9S,10S,13R,14S,17R)-3-(2-hydroxy-2-methylpropyl)-10,13-dimethylhexadecahydro-1H-cyclopenta[a]phenanthren-17-yl)pentanamido)propanoic acid, Mediator of RNA polymerase II transcription subunit 1, Vitamin D3 receptor | | Authors: | Kato, K, Numoto, N, Kagechika, H, Tanatani, A, Ito, N. | | Deposit date: | 2021-10-20 | | Release date: | 2022-03-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Lithocholic Acid Amides as Potent Vitamin D Receptor Agonists.
Biomolecules, 12, 2022
|
|
4NST
 
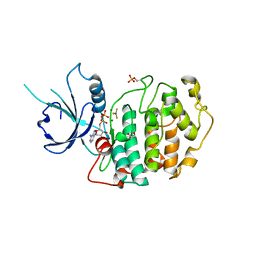 | | Crystal structure of human Cdk12/Cyclin K in complex with ADP-aluminum fluoride | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, ... | | Authors: | Boesken, C.A, Farnung, L, Anand, K, Geyer, M. | | Deposit date: | 2013-11-29 | | Release date: | 2014-03-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The structure and substrate specificity of human Cdk12/Cyclin K.
Nat Commun, 5, 2014
|
|
1B8I
 
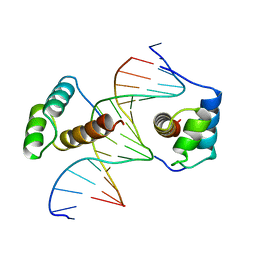 | | STRUCTURE OF THE HOMEOTIC UBX/EXD/DNA TERNARY COMPLEX | | Descriptor: | DNA (5'-D(*AP*CP*GP*TP*GP*AP*TP*TP*TP*AP*TP*GP*GP*CP*G)-3'), DNA (5'-D(*GP*TP*CP*GP*CP*CP*AP*TP*AP*AP*AP*TP*CP*AP*C)-3'), PROTEIN (HOMEOBOX PROTEIN EXTRADENTICLE), ... | | Authors: | Passner, J.M, Ryoo, H.-D, Shen, L, Mann, R.S, Aggarwal, A.K. | | Deposit date: | 1999-02-01 | | Release date: | 1999-04-12 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of a DNA-bound Ultrabithorax-Extradenticle homeodomain complex.
Nature, 397, 1999
|
|
1DRZ
 
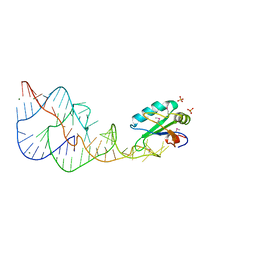 | | U1A SPLICEOSOMAL PROTEIN/HEPATITIS DELTA VIRUS GENOMIC RIBOZYME COMPLEX | | Descriptor: | MAGNESIUM ION, PROTEIN (U1 SMALL RIBONUCLEOPROTEIN A), RNA (HEPATITIS DELTA VIRUS GENOMIC RIBOZYME), ... | | Authors: | Ferre-D'Amare, A.R, Zhou, K, Doudna, J.A. | | Deposit date: | 1998-09-01 | | Release date: | 1999-02-16 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of a hepatitis delta virus ribozyme.
Nature, 395, 1998
|
|
2KIQ
 
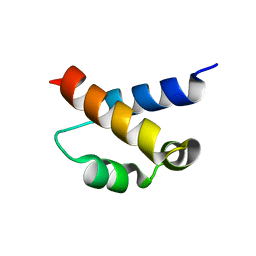 | | Solution structure of the FF Domain 2 of human transcription elongation factor CA150 | | Descriptor: | Transcription elongation regulator 1 | | Authors: | Zeng, J, Boyles, J, Tripathy, C, Yan, A, Zhou, P, Donald, B.R. | | Deposit date: | 2009-05-07 | | Release date: | 2009-07-28 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | High-resolution protein structure determination starting with a global fold calculated from exact solutions to the RDC equations.
J.Biomol.Nmr, 45, 2009
|
|
1E8S
 
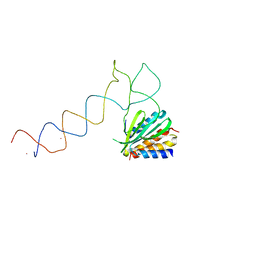 | | Alu domain of the mammalian SRP (potential Alu retroposition intermediate) | | Descriptor: | 7SL RNA, 88-MER, EUROPIUM (III) ION, ... | | Authors: | Weichenrieder, O, Wild, K, Strub, K, Cusack, S. | | Deposit date: | 2000-09-29 | | Release date: | 2000-11-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Structure and Assembly of the Alu Domain of the Mammalian Signal Recognition Particle
Nature, 408, 2000
|
|
6Q3C
 
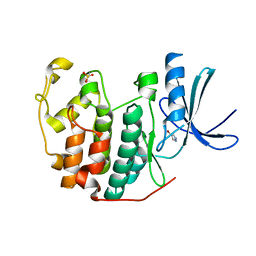 | | CDK2 in complex with FragLite1 | | Descriptor: | 4-bromo-1H-pyrazole, Cyclin-dependent kinase 2 | | Authors: | Wood, D.J, Martin, M.P, Noble, M.E.M. | | Deposit date: | 2018-12-04 | | Release date: | 2019-03-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | FragLites-Minimal, Halogenated Fragments Displaying Pharmacophore Doublets. An Efficient Approach to Druggability Assessment and Hit Generation.
J.Med.Chem., 62, 2019
|
|
6Q3B
 
 | | CDK2 in complex with FragLite2 | | Descriptor: | 4-IODOPYRAZOLE, Cyclin-dependent kinase 2 | | Authors: | Wood, D.J, Martin, M.P, Noble, M.E.M. | | Deposit date: | 2018-12-04 | | Release date: | 2019-03-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.11 Å) | | Cite: | FragLites-Minimal, Halogenated Fragments Displaying Pharmacophore Doublets. An Efficient Approach to Druggability Assessment and Hit Generation.
J.Med.Chem., 62, 2019
|
|
6Q49
 
 | | CDK2 in complex with FragLite6 | | Descriptor: | 4-bromanyl-1~{H}-pyridin-2-one, Cyclin-dependent kinase 2, DIMETHYL SULFOXIDE | | Authors: | Wood, D.J, Martin, M.P, Noble, M.E.M. | | Deposit date: | 2018-12-05 | | Release date: | 2019-03-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | FragLites-Minimal, Halogenated Fragments Displaying Pharmacophore Doublets. An Efficient Approach to Druggability Assessment and Hit Generation.
J.Med.Chem., 62, 2019
|
|
6Q4G
 
 | | CDK2 in complex with FragLite37 | | Descriptor: | 2-[3-(2-azanyl-9~{H}-purin-6-yl)phenyl]ethanoic acid, Cyclin-dependent kinase 2 | | Authors: | Wood, D.J, Martin, M.P, Noble, M.E.M. | | Deposit date: | 2018-12-05 | | Release date: | 2019-03-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (0.98 Å) | | Cite: | FragLites-Minimal, Halogenated Fragments Displaying Pharmacophore Doublets. An Efficient Approach to Druggability Assessment and Hit Generation.
J.Med.Chem., 62, 2019
|
|
6Q48
 
 | | CDK2 in complex with FragLite7 | | Descriptor: | 4-iodanyl-3~{H}-pyridin-2-one, Cyclin-dependent kinase 2, DIMETHYL SULFOXIDE | | Authors: | Wood, D.J, Martin, M.P, Noble, M.E.M. | | Deposit date: | 2018-12-05 | | Release date: | 2019-03-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.03 Å) | | Cite: | FragLites-Minimal, Halogenated Fragments Displaying Pharmacophore Doublets. An Efficient Approach to Druggability Assessment and Hit Generation.
J.Med.Chem., 62, 2019
|
|
