6QDZ
 
 | | P38 alpha complex with AR117045 | | Descriptor: | 1-[5-~{tert}-butyl-2-(4-methylphenyl)pyrazol-3-yl]-3-[(1~{S},4~{S})-4-[(3-propan-2-yl-[1,2,4]triazolo[4,3-a]pyridin-6-yl)oxy]-1,2,3,4-tetrahydronaphthalen-1-yl]urea, 2-fluoro-4-[4-(4-fluorophenyl)-1H-pyrazol-3-yl]pyridine, Mitogen-activated protein kinase 14 | | Authors: | Brown, D.G, Hurley, C, Irving, S.L. | | Deposit date: | 2019-01-03 | | Release date: | 2020-01-29 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | P38 alpha complex with AR117045
To Be Published
|
|
7K11
 
 | |
8WQC
 
 | | cryo-EM structure of neddylated CUL2-RBX1-ELOB-ELOC-FEM1B bound with the C-degron of CDK5R1 | | Descriptor: | Cullin-2, E3 ubiquitin-protein ligase RBX1, N-terminally processed, ... | | Authors: | Chen, X, Zhang, K, Xu, C. | | Deposit date: | 2023-10-11 | | Release date: | 2024-04-03 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.54 Å) | | Cite: | Mechanism of Psi-Pro/C-degron recognition by the CRL2 FEM1B ubiquitin ligase.
Nat Commun, 15, 2024
|
|
8WQG
 
 | | cryo-EM structure of neddylated CUL2-RBX1-ELOB-ELOC-FEM1B bound with the C-degron of CCDC89 (conformation 1) | | Descriptor: | Cullin-2, E3 ubiquitin-protein ligase RBX1, N-terminally processed, ... | | Authors: | Chen, X, Zhang, K, Xu, C. | | Deposit date: | 2023-10-11 | | Release date: | 2024-04-03 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (4.09 Å) | | Cite: | Mechanism of Psi-Pro/C-degron recognition by the CRL2 FEM1B ubiquitin ligase.
Nat Commun, 15, 2024
|
|
7K78
 
 | | antibody and nucleosome complex | | Descriptor: | Cse4, DNA (136-MER), Histone H2A.1, ... | | Authors: | Ruifang, G, Yawen, B. | | Deposit date: | 2020-09-22 | | Release date: | 2021-03-31 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural and dynamic mechanisms of CBF3-guided centromeric nucleosome formation.
Nat Commun, 12, 2021
|
|
6V8O
 
 | | RSC core | | Descriptor: | Chromatin structure-remodeling complex protein RSC14, Chromatin structure-remodeling complex protein RSC3, Chromatin structure-remodeling complex protein RSC30, ... | | Authors: | Patel, A.B, Moore, C.M, Greber, B.J, Nogales, E. | | Deposit date: | 2019-12-11 | | Release date: | 2020-01-15 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (3.07 Å) | | Cite: | Architecture of the chromatin remodeler RSC and insights into its nucleosome engagement.
Elife, 8, 2019
|
|
5O8V
 
 | |
6QE1
 
 | | P38 alpha complex with AR117046 | | Descriptor: | 1-[5-~{tert}-butyl-2-(4-methylphenyl)pyrazol-3-yl]-3-[(1~{S},4~{R})-4-[(3-propan-2-yl-[1,2,4]triazolo[4,3-a]pyridin-6-yl)oxy]-1,2,3,4-tetrahydronaphthalen-1-yl]urea, 2-fluoro-4-[4-(4-fluorophenyl)-1H-pyrazol-3-yl]pyridine, Mitogen-activated protein kinase 14 | | Authors: | Brown, D.G, Hurley, C, Irving, S.L. | | Deposit date: | 2019-01-03 | | Release date: | 2020-01-29 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | P38 alpha complex with AR117045 and AR117046
To Be Published
|
|
5XYX
 
 | | The structure of p38 alpha in complex with a triazol inhibitor | | Descriptor: | Mitogen-activated protein kinase 14, N-(2-chloro-6-fluorobenzyl)-5-(furan-2-yl)-2H-1,2,4-triazol-3-amine | | Authors: | Wang, Y.L, Sun, Y.Z, Cao, R, Liu, D, Li, L, Qi, X.B, Huang, N. | | Deposit date: | 2017-07-11 | | Release date: | 2018-01-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | In Silico Identification of a Novel Hinge-Binding Scaffold for Kinase Inhibitor Discovery.
J. Med. Chem., 60, 2017
|
|
1UBB
 
 | | Crystal structure of rat HO-1 in complex with ferrous heme | | Descriptor: | Heme oxygenase 1, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Sugishima, M, Sakamoto, H, Higashimoto, Y, Noguchi, M, Fukuyama, K. | | Deposit date: | 2003-04-03 | | Release date: | 2003-09-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structures of Ferrous and CO-, CN(-)-, and NO-Bound Forms of Rat Heme Oxygenase-1 (HO-1) in Complex with Heme: Structural Implications for Discrimination between CO and O(2) in HO-1.
Biochemistry, 42, 2003
|
|
5XYY
 
 | | The structure of p38 alpha in complex with a triazol inhibitor | | Descriptor: | 3-(5-{[(2-chloro-6-fluorophenyl)methyl]amino}-4H-1,2,4-triazol-3-yl)phenol, Mitogen-activated protein kinase 14 | | Authors: | Wang, Y.L, Sun, Y.Z, Cao, R, Liu, D, Li, L, Qi, X.B, Huang, N. | | Deposit date: | 2017-07-11 | | Release date: | 2018-01-17 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | In Silico Identification of a Novel Hinge-Binding Scaffold for Kinase Inhibitor Discovery.
J. Med. Chem., 60, 2017
|
|
1U2V
 
 | | Crystal structure of Arp2/3 complex with bound ADP and calcium | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin-Related Protein 2, Actin-Related Protein 3, ... | | Authors: | Nolen, B.J, Littlefield, R.S, Pollard, T.D. | | Deposit date: | 2004-07-20 | | Release date: | 2004-11-09 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystal structures of actin-related protein 2/3 complex with bound ATP or ADP
Proc.Natl.Acad.Sci.Usa, 101, 2004
|
|
8GA8
 
 | | Structure of the yeast (HDAC) Rpd3L complex | | Descriptor: | Histone deacetylase RPD3, Transcriptional regulatory protein DEP1, Transcriptional regulatory protein PHO23, ... | | Authors: | Patel, A.B, Radhakrishnan, I, He, Y. | | Deposit date: | 2023-02-22 | | Release date: | 2023-07-12 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-EM structure of the Saccharomyces cerevisiae Rpd3L histone deacetylase complex.
Nat Commun, 14, 2023
|
|
8CG3
 
 | |
1A9U
 
 | | THE COMPLEX STRUCTURE OF THE MAP KINASE P38/SB203580 | | Descriptor: | 4-[5-(4-FLUORO-PHENYL)-2-(4-METHANESULFINYL-PHENYL)-3H-IMIDAZOL-4-YL]-PYRIDINE, MAP KINASE P38 | | Authors: | Wang, Z, Canagarajah, B, Boehm, J.C, Kassis, S, Cobb, M.H, Young, P.R, Abdel-Meguid, S, Adams, J.L, Goldsmith, E.J. | | Deposit date: | 1998-04-10 | | Release date: | 1999-04-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis of inhibitor selectivity in MAP kinases.
Structure, 6, 1998
|
|
7TDZ
 
 | |
6SFO
 
 | | MAPK14 with bound inhibitor SR-318 | | Descriptor: | 5-azanyl-~{N}-[[4-(3-cyclohexylpropylcarbamoyl)phenyl]methyl]-1-phenyl-pyrazole-4-carboxamide, GLYCEROL, Mitogen-activated protein kinase 14, ... | | Authors: | Schroeder, M, Edwards, A.M, Arrowsmith, C.H, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-08-01 | | Release date: | 2019-09-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | MAPK14 with bound inhibitor SR-318
To Be Published
|
|
9D8P
 
 | | Focused map of Cryo-EM structure of Ubiquitin C-degron bound to KLHDC10-EloB/C | | Descriptor: | Elongin-B, Elongin-C, Kelch domain-containing protein 10, ... | | Authors: | Chittori, S, Scott, D.C, Schulman, B.A. | | Deposit date: | 2024-08-19 | | Release date: | 2024-11-20 | | Last modified: | 2025-02-05 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis for C-degron selectivity across KLHDCX family E3 ubiquitin ligases.
Nat Commun, 15, 2024
|
|
9DDE
 
 | | ncPRC1RYBP bound to H2AK119Ub/H1.4 chromatosome | | Descriptor: | DNA (187-MER), E3 ubiquitin-protein ligase RING2, Histone H1.4, ... | | Authors: | Godinez-Lopez, V, Valencia-Sanchez, M.I, Armache, J.P, Armache, K.-J. | | Deposit date: | 2024-08-28 | | Release date: | 2024-11-20 | | Last modified: | 2025-01-15 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Read-write mechanisms of H2A ubiquitination by Polycomb repressive complex 1.
Nature, 636, 2024
|
|
5VHF
 
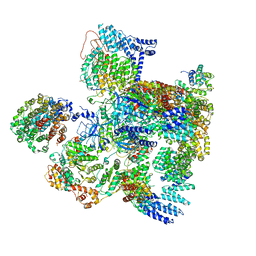 | | Conformational Landscape of the p28-Bound Human Proteasome Regulatory Particle | | Descriptor: | 26S proteasome complex subunit SEM1, 26S proteasome non-ATPase regulatory subunit 1, 26S proteasome non-ATPase regulatory subunit 10, ... | | Authors: | Lu, Y, Wu, J, Dong, Y, Chen, S, Sun, S, Ma, Y.B, Ouyang, Q, Finley, D, Kirschner, M.W, Mao, Y. | | Deposit date: | 2017-04-13 | | Release date: | 2017-08-23 | | Last modified: | 2023-08-16 | | Method: | ELECTRON MICROSCOPY (5.7 Å) | | Cite: | Conformational Landscape of the p28-Bound Human Proteasome Regulatory Particle.
Mol. Cell, 67, 2017
|
|
7UX2
 
 | |
3MHS
 
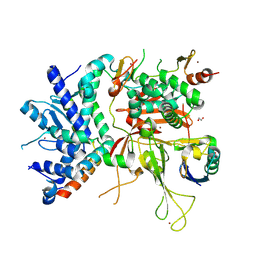 | | Structure of the SAGA Ubp8/Sgf11/Sus1/Sgf73 DUB module bound to ubiquitin aldehyde | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Protein SUS1, ... | | Authors: | Samara, N.L, Datta, A.B, Berndsen, C.E, Zhang, X, Yao, T, Cohen, R.E, Wolberger, C. | | Deposit date: | 2010-04-08 | | Release date: | 2010-04-21 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structural insights into the assembly and function of the SAGA deubiquitinating module.
Science, 328, 2010
|
|
6ANL
 
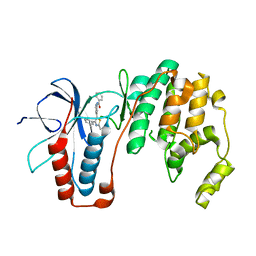 | | Structure-based Design, Synthesis, and Biological Evaluation of Imidazo[1,2-b]pyridazine-based p38 MAP Kinase Inhibitors | | Descriptor: | Mitogen-activated protein kinase 14, TAK-715 | | Authors: | Snell, G.P, Okada, K, Bragstad, K, Sang, B.-C. | | Deposit date: | 2017-08-14 | | Release date: | 2018-01-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-based design, synthesis, and biological evaluation of imidazo[1,2-b]pyridazine-based p38 MAP kinase inhibitors.
Bioorg. Med. Chem., 26, 2018
|
|
3MHH
 
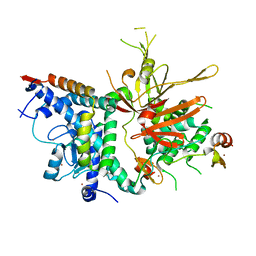 | | Structure of the SAGA Ubp8/Sgf11/Sus1/Sgf73 DUB module | | Descriptor: | Protein SUS1, SAGA-associated factor 11, SAGA-associated factor 73, ... | | Authors: | Samara, N.L, Datta, A.B, Berndsen, C.E, Zhang, X, Yao, T, Cohen, R.E, Wolberger, C. | | Deposit date: | 2010-04-08 | | Release date: | 2010-04-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structural insights into the assembly and function of the SAGA deubiquitinating module.
Science, 328, 2010
|
|
6GMR
 
 | | pVHL:EloB:EloC in complex with (4-(1H-pyrrol-1-yl)phenyl)methanol | | Descriptor: | (4-pyrrol-1-ylphenyl)methanol, Elongin-B, Elongin-C, ... | | Authors: | Van Molle, I, Lucas, X, Ciulli, A. | | Deposit date: | 2018-05-28 | | Release date: | 2018-08-08 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Surface Probing by Fragment-Based Screening and Computational Methods Identifies Ligandable Pockets on the von Hippel-Lindau (VHL) E3 Ubiquitin Ligase.
J. Med. Chem., 61, 2018
|
|
