2BH2
 
 | | Crystal Structure of E. coli 5-methyluridine methyltransferase RumA in complex with ribosomal RNA substrate and S-adenosylhomocysteine. | | Descriptor: | 23S RIBOSOMAL RNA 1932-1968, 23S RRNA (URACIL-5-)-METHYLTRANSFERASE RUMA, IRON/SULFUR CLUSTER, ... | | Authors: | Lee, T.T, Agarwalla, S, Stroud, R.M. | | Deposit date: | 2005-01-06 | | Release date: | 2005-03-30 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | A Unique RNA Fold in the Ruma-RNA-Cofactor Ternary Complex Contributes to Substrate Selectivity and Enzymatic Function
Cell(Cambridge,Mass.), 120, 2005
|
|
2BSX
 
 | | Crystal structure of the Plasmodium falciparum purine nucleoside phosphorylase complexed with inosine | | Descriptor: | INOSINE, PURINE NUCLEOSIDE PHOSPHORYLASE | | Authors: | Schnick, C, Brzozowski, A.M, Dodson, E.J, Murshudov, G.N, Brannigan, J.A, Wilkinson, A.J. | | Deposit date: | 2005-05-24 | | Release date: | 2005-08-18 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of Plasmodium Falciparum Purine Nucleoside Phosphorylase Complexed with Sulfate and its Natural Substrate Inosine
Acta Crystallogr.,Sect.D, 61, 2005
|
|
6CSM
 
 | | Crystal structure of the natural light-gated anion channel GtACR1 | | Descriptor: | GtACR1, OLEIC ACID, RETINAL | | Authors: | Kato, H.E, Kim, Y, Yamashita, K, Kobilka, B.K, Deisseroth, K. | | Deposit date: | 2018-03-21 | | Release date: | 2018-09-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural mechanisms of selectivity and gating in anion channelrhodopsins.
Nature, 561, 2018
|
|
2BJN
 
 | | X-ray Structure of human TPC6 | | Descriptor: | GLYCEROL, SULFATE ION, TRAFFICKING PROTEIN PARTICLE COMPLEX SUBUNIT 6B | | Authors: | Kummel, D, Mueller, J.J, Roske, Y, Misselwitz, R, Bussow, K, Heinemann, U. | | Deposit date: | 2005-02-04 | | Release date: | 2005-07-20 | | Last modified: | 2025-10-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Structure of the Trapp Subunit Tpc6 Suggests a Model for a Trapp Subcomplex.
Embo Rep., 6, 2005
|
|
2BIK
 
 | | Human Pim1 phosphorylated on Ser261 | | Descriptor: | 3-{1-[3-(DIMETHYLAMINO)PROPYL]-1H-INDOL-3-YL}-4-(1H-INDOL-3-YL)-1H-PYRROLE-2,5-DIONE, CHLORIDE ION, PROTO-ONCOGENE SERINE/THREONINE-PROTEIN KINASE PIM-1 | | Authors: | Knapp, S, Debreczeni, J, Bullock, A, von Delft, F, Sundstrom, M, Arrowsmith, C, Edwards, A, Guo, K. | | Deposit date: | 2005-01-22 | | Release date: | 2005-02-07 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Human Pim1 Phosphorylated on Ser261
To be Published
|
|
2BJQ
 
 | | Crystal structure of the nematode sperm cell motility protein MFP2 | | Descriptor: | MFP2A | | Authors: | Grant, R.P, Buttery, S.M, Ekman, G.C, Roberts, T.M, Stewart, M. | | Deposit date: | 2005-02-07 | | Release date: | 2005-03-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure of Mfp2 and its Function in Enhancing Msp Polymerization in Ascaris Sperm Amoeboid Motility
J.Mol.Biol., 347, 2005
|
|
2BK4
 
 | | Human Monoamine Oxidase B: I199F mutant in complex with rasagiline | | Descriptor: | (1R)-N-(prop-2-en-1-yl)-2,3-dihydro-1H-inden-1-amine, AMINE OXIDASE [FLAVIN-CONTAINING] B, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Binda, C, Edmondson, D.E, Mattevi, A, Hubalek, F, Khalil, A, Li, M, Castagnoli, N. | | Deposit date: | 2005-02-10 | | Release date: | 2005-02-14 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Demonstration of Isoleucine 199 as a Structural Determinant for the Selective Inhibition of Human Monoamine Oxidase B by Specific Reversible Inhibitors
J.Biol.Chem., 280, 2005
|
|
2BOP
 
 | | CRYSTAL STRUCTURE AT 1.7 ANGSTROMS OF THE BOVINE PAPILLOMAVIRUS-1 E2 DNA-BINDING DOMAIN BOUND TO ITS DNA TARGET | | Descriptor: | DNA (5'-D(*CP*CP*GP*AP*CP*CP*GP*AP*CP*GP*TP*CP*GP*GP*TP*CP*G )-3'), PROTEIN (E2), YTTERBIUM (III) ION | | Authors: | Hegde, R.S, Grossman, S.R, Laimins, L.A, Sigler, P.B. | | Deposit date: | 1994-01-13 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure at 1.7 A of the bovine papillomavirus-1 E2 DNA-binding domain bound to its DNA target.
Nature, 359, 1992
|
|
6CC6
 
 | |
6CCN
 
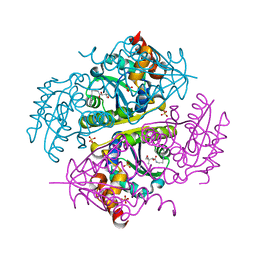 | | Crystal structure of E.coli Phosphopantetheine Adenylyltransferase (PPAT/CoaD) in complex with (R)-2,4-dihydroxy-N-(2-(4-hydroxy-1H-benzo[d]imidazol-2-yl)ethyl)-3,3-dimethylbutanamide | | Descriptor: | (2R)-2,4-dihydroxy-N-[2-(7-hydroxy-1H-benzimidazol-2-yl)ethyl]-3,3-dimethylbutanamide, Phosphopantetheine adenylyltransferase, SULFATE ION | | Authors: | Mamo, M, Appleton, B.A. | | Deposit date: | 2018-02-07 | | Release date: | 2018-03-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Fragment-Based Drug Discovery of Inhibitors of Phosphopantetheine Adenylyltransferase from Gram-Negative Bacteria.
J. Med. Chem., 61, 2018
|
|
2C2Z
 
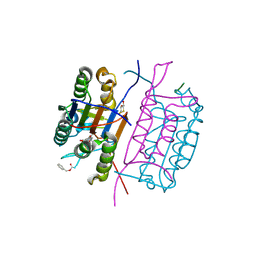 | | Crystal structure of caspase-8 in complex with aza-peptide Michael acceptor inhibitor | | Descriptor: | AZA-PEPTIDE INHIBITOR (5S, 8R, 11S)-8-(2-CARBOXYETHYL) -14-[4-(3,4-DIHYDROQUINOLIN-1(2H)-YL)-4-OXOBUTANOYL] -11-[(1R)-1-HYDROXYETHYL]-5-(2-METHYLPROPYL)-3,6,9,12-TETRAOXO -1-PHENYL-2-OXA-4,7,10,13,14-PENTAAZAHEXADECAN-16-OIC ACID, ... | | Authors: | Ganesan, R, Jelakovic, S, Ekici, O.D, Li, Z.Z, James, K.E, Asgian, J.L, Campbell, A.J, Mikolajczyk, J, Salvesen, G.S, Powers, J.C, Gruetter, M.G. | | Deposit date: | 2005-10-02 | | Release date: | 2006-09-20 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Design, Synthesis, and Evaluation of Aza-Peptide Michael Acceptors as Selective and Potent Inhibitors of Caspases-2, -3, -6, -7, -8, -9, and - 10.
J.Med.Chem., 49, 2006
|
|
2BND
 
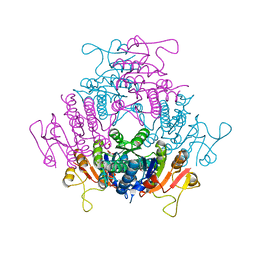 | | The structure of E. coli UMP kinase in complex with UDP | | Descriptor: | GLYCEROL, PYROPHOSPHATE 2-, URIDINE-5'-DIPHOSPHATE, ... | | Authors: | Briozzo, P, Evrin, C, Meyer, P, Assairi, L, Joly, N, Barzu, O, Gilles, A.M. | | Deposit date: | 2005-03-23 | | Release date: | 2005-04-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of Escherichia Coli Ump Kinase Differs from that of Other Nucleoside Monophosphate Kinases and Sheds New Light on Enzyme Regulation.
J.Biol.Chem., 280, 2005
|
|
6CTQ
 
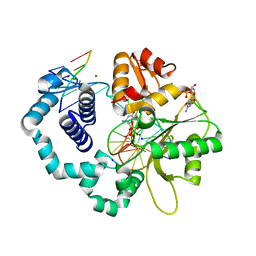 | |
2BXD
 
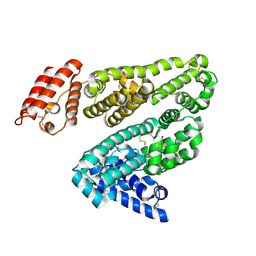 | | Human serum albumin complexed with warfarin | | Descriptor: | R-WARFARIN, SERUM ALBUMIN | | Authors: | Ghuman, J, Zunszain, P.A, Petitpas, I, Bhattacharya, A.A, Curry, S. | | Deposit date: | 2005-07-26 | | Release date: | 2005-09-22 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Structural Basis of the Drug-Binding Specificity of Human Serum Albumin.
J.Mol.Biol., 353, 2005
|
|
2BRZ
 
 | | SOLUTION NMR STRUCTURE OF THE SWEET PROTEIN BRAZZEIN, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | BRAZZEIN | | Authors: | Caldwell, J.E, Abildgaard, F, Dzakula, Z, Ming, D, Hellekant, G, Markley, J.L. | | Deposit date: | 1998-04-30 | | Release date: | 1998-07-01 | | Last modified: | 2024-11-20 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the thermostable sweet-tasting protein brazzein.
Nat.Struct.Biol., 5, 1998
|
|
6CCU
 
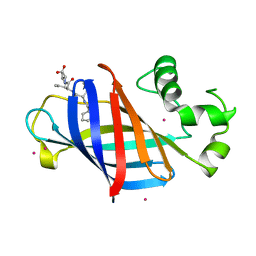 | | Complex between a GID4 fragment and a short peptide | | Descriptor: | Glucose-induced degradation protein 4 homolog, Short peptide, UNKNOWN ATOM OR ION | | Authors: | Dong, C, Tempel, W, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-02-07 | | Release date: | 2018-03-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Molecular basis of GID4-mediated recognition of degrons for the Pro/N-end rule pathway.
Nat. Chem. Biol., 14, 2018
|
|
6CD9
 
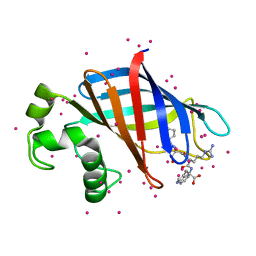 | | GID4 in complex with a peptide | | Descriptor: | Glucose-induced degradation protein 4 homolog, Tetrapeptide PSRW, UNKNOWN ATOM OR ION | | Authors: | Dong, C, Tempel, W, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-02-08 | | Release date: | 2018-03-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Molecular basis of GID4-mediated recognition of degrons for the Pro/N-end rule pathway.
Nat. Chem. Biol., 14, 2018
|
|
2BJW
 
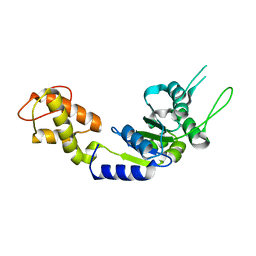 | | PspF AAA domain | | Descriptor: | PSP OPERON TRANSCRIPTIONAL ACTIVATOR | | Authors: | Rappas, M, Schumacher, J, Beuron, F, Niwa, H, Bordes, P, Wigneshweraraj, S, Keetch, C.A, Robinson, C.V, Buck, M, Zhang, X. | | Deposit date: | 2005-02-08 | | Release date: | 2005-03-31 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural Insights Into the Activity of Enhancer-Binding Proteins
Science, 307, 2005
|
|
6CU6
 
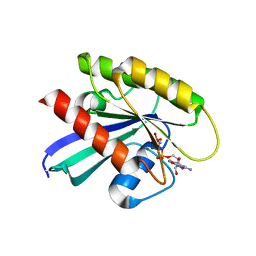 | |
2C5X
 
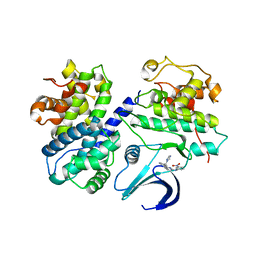 | | Differential Binding Of Inhibitors To Active And Inactive Cdk2 Provides Insights For Drug Design | | Descriptor: | CELL DIVISION PROTEIN KINASE 2, CYCLIN A2, HYDROXY(OXO)(3-{[(2Z)-4-[3-(1H-1,2,4-TRIAZOL-1-YLMETHYL)PHENYL]PYRIMIDIN-2(5H)-YLIDENE]AMINO}PHENYL)AMMONIUM | | Authors: | Kontopidis, G, Mcinnes, C, Pandalaneni, S.R, Mcnae, I, Gibson, D, Mezna, M, Thomas, M, Wood, G, Wang, S, Walkinshaw, M.D, Fischer, P.M. | | Deposit date: | 2005-11-03 | | Release date: | 2006-03-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Differential Binding of Inhibitors to Active and Inactive Cdk2 Provides Insights for Drug Design.
Chem.Biol., 13, 2006
|
|
2BDV
 
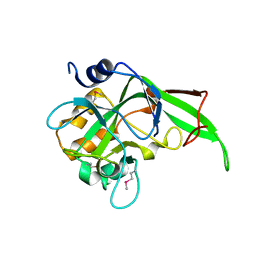 | | X-Ray Crystal Structure of Phage-related Protein BB2244 from Bordetella bronchiseptica. Northeast Structural Genomics Consortium Target BoR24. | | Descriptor: | phage-related conserved hypothetical protein, BB2244 | | Authors: | Forouhar, F, Abashidze, M, Benach, J, Jayaraman, S, Janjua, H, Cooper, B, Xiao, R, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2005-10-20 | | Release date: | 2005-11-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of the phage-related conserved hypothetical protein BB2244 from Bordetella bronchiseptica, Northeast Structural Genomics Target BoR24
To be Published
|
|
6CDA
 
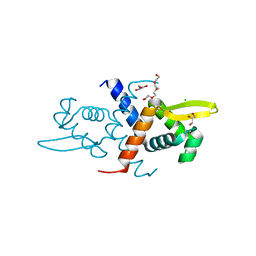 | | Crystal structure of L34A CzrA in the Zn(II)bound state | | Descriptor: | ArsR family transcriptional regulator, CHLORIDE ION, GLYCEROL, ... | | Authors: | Capdevila, D.A, Gonzalez-Gutierrez, G, Giedroc, D.P. | | Deposit date: | 2018-02-08 | | Release date: | 2018-07-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Functional Role of Solvent Entropy and Conformational Entropy of Metal Binding in a Dynamically Driven Allosteric System.
J. Am. Chem. Soc., 140, 2018
|
|
2BU6
 
 | | crystal structures of human pyruvate dehydrogenase kinase 2 containing physiological and synthetic ligands | | Descriptor: | (N-{4-[(ETHYLANILINO)SULFONYL]-2-METHYLPHENYL}-3,3,3-TRIFLUORO-2-HYDROXY-2-METHYLPROPANAMIDE, PYRUVATE DEHYDROGENASE KINASE ISOENZYME 2 | | Authors: | Knoechel, T.R, Tucker, A.D, Robinson, C.M, Phillips, C, Taylor, W, Bungay, P.J, Kasten, S.A, Roche, T.E, Brown, D.G. | | Deposit date: | 2005-06-08 | | Release date: | 2006-02-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Regulatory Roles of the N-Terminal Domain Based on Crystal Structures of Human Pyruvate Dehydrogenase Kinase 2 Containing Physiological and Synthetic Ligands.
Biochemistry, 45, 2006
|
|
6CVT
 
 | | Human Aprataxin (Aptx) V263G bound to RNA-DNA, AMP and Zn product complex | | Descriptor: | ADENOSINE MONOPHOSPHATE, Aprataxin, DNA (5'-D(*GP*AP*AP*TP*CP*AP*TP*AP*AP*C)-3'), ... | | Authors: | Schellenberg, M.J, Tumbale, P.S, Williams, R.S. | | Deposit date: | 2018-03-28 | | Release date: | 2018-07-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.941 Å) | | Cite: | Mechanism of APTX nicked DNA sensing and pleiotropic inactivation in neurodegenerative disease.
EMBO J., 37, 2018
|
|
2BE1
 
 | | Structure of the compact lumenal domain of yeast Ire1 | | Descriptor: | Serine/threonine-protein kinase/endoribonuclease IRE1, peptide | | Authors: | Credle, J.J, Finer-Moore, J.S, Papa, F.R, Stroud, R.M, Walter, P. | | Deposit date: | 2005-10-21 | | Release date: | 2005-12-13 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.983 Å) | | Cite: | Inaugural Article: On the mechanism of sensing unfolded protein in the endoplasmic reticulum
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
