7MKP
 
 | | Escherichia coli RNA polymerase core enzyme | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Qayyum, M.Z, Murakami, K.S. | | Deposit date: | 2021-04-26 | | Release date: | 2021-06-23 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.41 Å) | | Cite: | Structural basis of RNA polymerase recycling by the Swi2/Snf2 family of ATPase RapA in Escherichia coli.
J.Biol.Chem., 297, 2021
|
|
7ZF2
 
 | | Protomeric substructure from an octameric assembly of M. tuberculosis RNA polymerase in complex with sigma-b initiation factor | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Trapani, S, Bron, P, Lai Kee Him, J, Brodolin, K, Morichaud, Z, Vishwakarma, R. | | Deposit date: | 2022-03-31 | | Release date: | 2023-02-08 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.86 Å) | | Cite: | Structural basis of the mycobacterial stress-response RNA polymerase auto-inhibition via oligomerization.
Nat Commun, 14, 2023
|
|
8TO6
 
 | | Escherichia coli RNA polymerase unwinding intermediate (I1d) at the lambda PR promoter | | Descriptor: | (3R,5S,7R,8R,9S,10S,12S,13R,14S,17R)-10,13-dimethyl-17-[(2R)-pentan-2-yl]-2,3,4,5,6,7,8,9,11,12,14,15,16,17-tetradecahydro-1H-cyclopenta[a]phenanthrene-3,7,12-triol, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Darst, S.A, Saecker, R.M, Mueller, A.U. | | Deposit date: | 2023-08-02 | | Release date: | 2024-07-03 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Early intermediates in bacterial RNA polymerase promoter melting visualized by time-resolved cryo-electron microscopy.
Nat.Struct.Mol.Biol., 2024
|
|
1UVK
 
 | | The structural basis for RNA specificity and Ca2 inhibition of an RNA-dependent RNA polymerase phi6p2 dead-end complex | | Descriptor: | GUANOSINE-5'-MONOPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Salgado, P.S, Makeyev, E.V, Butcher, S, Bamford, D, Stuart, D.I, Grimes, J.M. | | Deposit date: | 2004-01-21 | | Release date: | 2004-02-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | The structural basis for RNA specificity and Ca2+ inhibition of an RNA-dependent RNA polymerase.
Structure, 12, 2004
|
|
3QID
 
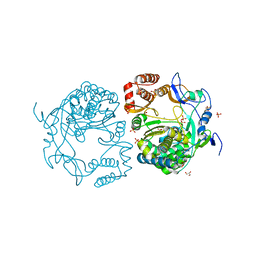 | | Crystal structures and functional analysis of murine norovirus RNA-dependent RNA polymerase | | Descriptor: | GLYCEROL, MANGANESE (III) ION, RNA dependent RNA polymerase, ... | | Authors: | Kim, K.H, Intekhab, A, Lee, J.H. | | Deposit date: | 2011-01-27 | | Release date: | 2011-12-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of murine norovirus-1 RNA-dependent RNA polymerase.
J.Gen.Virol., 92, 2011
|
|
3MMP
 
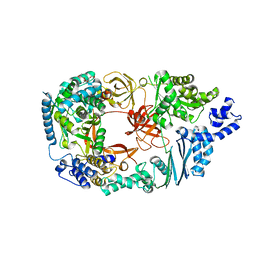 | | Structure of the Qb replicase, an RNA-dependent RNA polymerase consisting of viral and host proteins | | Descriptor: | (2S)-1-[3-{[(2R)-2-hydroxypropyl]oxy}-2,2-bis({[(2R)-2-hydroxypropyl]oxy}methyl)propoxy]propan-2-ol, Elongation factor Tu 2, Elongation factor Ts, ... | | Authors: | Kidmose, R.T, Vasiliev, N.N, Chetverin, A.B, Knudsen, C.R, Andersen, G.R. | | Deposit date: | 2010-04-20 | | Release date: | 2010-06-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of the Qbeta replicase, an RNA-dependent RNA polymerase consisting of viral and host proteins.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3SPA
 
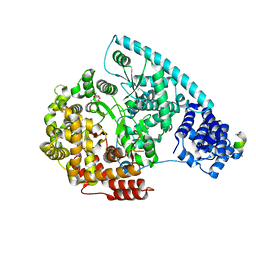 | | Crystal Structure of Human Mitochondrial RNA Polymerase | | Descriptor: | CHLORIDE ION, DNA-directed RNA polymerase, mitochondrial, ... | | Authors: | Ringel, R, Sologub, M, Morozov, Y.I, Litonin, D, Cramer, P, Temiakov, D. | | Deposit date: | 2011-07-01 | | Release date: | 2011-09-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of human mitochondrial RNA polymerase
Nature, 478, 2011
|
|
2A6E
 
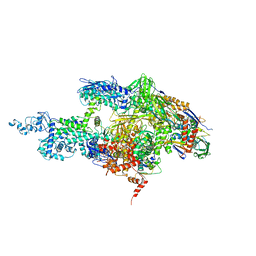 | | Crystal structure of the T. Thermophilus RNA polymerase holoenzyme | | Descriptor: | DNA-directed RNA polymerase alpha chain, DNA-directed RNA polymerase beta chain, DNA-directed RNA polymerase beta' chain, ... | | Authors: | Artsimovitch, I, Vassylyeva, M.N, Svetlov, D, Svetlov, V, Perederina, A, Igarashi, N, Matsugaki, N, Wakatsuki, S, Tahirov, T.H, Vassylyev, D.G, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-07-02 | | Release date: | 2005-09-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Allosteric modulation of the RNA polymerase catalytic reaction is an essential component of transcription control by rifamycins.
Cell(Cambridge,Mass.), 122, 2005
|
|
6PO2
 
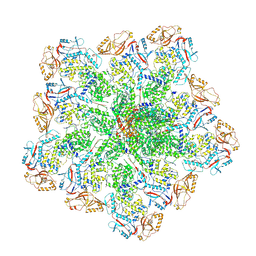 | | In situ structure of BTV RNA-dependent RNA polymerase in BTV core | | Descriptor: | Inner core structural protein VP3, RNA-directed RNA polymerase | | Authors: | He, Y, Shivakoti, S, Ding, K, Cui, Y, Roy, P, Zhou, Z.H. | | Deposit date: | 2019-07-03 | | Release date: | 2019-08-07 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | In situ structures of RNA-dependent RNA polymerase inside bluetongue virus before and after uncoating.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6PNS
 
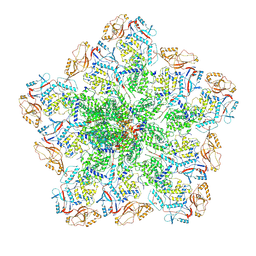 | | In situ structure of BTV RNA-dependent RNA polymerase in BTV virion | | Descriptor: | Inner core structural protein VP3, RNA-directed RNA polymerase | | Authors: | He, Y, Shivakoti, S, Ding, K, Cui, Y, Roy, P, Zhou, Z.H. | | Deposit date: | 2019-07-03 | | Release date: | 2019-08-07 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | In situ structures of RNA-dependent RNA polymerase inside bluetongue virus before and after uncoating.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
2CW0
 
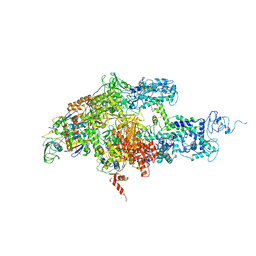 | | Crystal structure of Thermus thermophilus RNA polymerase holoenzyme at 3.3 angstroms resolution | | Descriptor: | DNA-directed RNA polymerase alpha chain, DNA-directed RNA polymerase beta chain, DNA-directed RNA polymerase beta' chain, ... | | Authors: | Tuske, S, Sarafianos, S.G, Wang, X, Hudson, B, Sineva, E, Mukhopadhyay, J, Birktoft, J.J, Leroy, O, Ismail, S, Clark Jr, A.D, Dharia, C, Napoli, A, Laptenko, O, Lee, J, Borukhov, S, Ebright, R.H, Arnold, E. | | Deposit date: | 2005-06-15 | | Release date: | 2005-09-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Inhibition of bacterial RNA polymerase by streptolydigin: stabilization of a straight-bridge-helix active-center conformation
Cell(Cambridge,Mass.), 122, 2005
|
|
2EC0
 
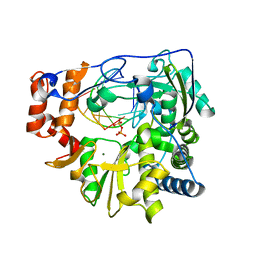 | | RNA-dependent RNA polymerase of foot-and-mouth disease virus in complex with a template-primer RNA and ATP | | Descriptor: | 5'-R(*GP*GP*GP*CP*CP*CP*A)-3', 5'-R(P*AP*UP*GP*GP*GP*CP*CP*C)-3', MAGNESIUM ION, ... | | Authors: | Ferrer-Orta, C, Arias, A, Perez-Luque, R, Escarmis, C, Domingo, E, Verdaguer, N. | | Deposit date: | 2007-02-09 | | Release date: | 2007-06-26 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Sequential structures provide insights into the fidelity of RNA replication
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
5VT0
 
 | |
3SFU
 
 | | crystal structure of murine norovirus RNA dependent RNA polymerase in complex with ribavirin | | Descriptor: | 1-(beta-D-ribofuranosyl)-1H-1,2,4-triazole-3-carboxamide, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Kim, K.H, Alam, I. | | Deposit date: | 2011-06-14 | | Release date: | 2012-05-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | Crystal structures of murine norovirus-1 RNA-dependent RNA polymerase in complex with 2-thiouridine or ribavirin.
Virology, 426, 2012
|
|
2A69
 
 | | Crystal structure of the T. Thermophilus RNA polymerase holoenzyme in complex with antibiotic rifapentin | | Descriptor: | DNA-directed RNA polymerase alpha chain, DNA-directed RNA polymerase beta chain, DNA-directed RNA polymerase beta' chain, ... | | Authors: | Artsimovitch, I, Vassylyeva, M.N, Svetlov, D, Svetlov, V, Perederina, A, Igarashi, N, Matsugaki, N, Wakatsuki, S, Tahirov, T.H, Vassylyev, D.G, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-07-02 | | Release date: | 2005-09-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Allosteric modulation of the RNA polymerase catalytic reaction is an essential component of transcription control by rifamycins.
Cell(Cambridge,Mass.), 122, 2005
|
|
2A68
 
 | | Crystal structure of the T. thermophilus RNA polymerase holoenzyme in complex with antibiotic rifabutin | | Descriptor: | DNA-directed RNA polymerase alpha chain, DNA-directed RNA polymerase beta chain, DNA-directed RNA polymerase beta' chain, ... | | Authors: | Artsimovitch, I, Vassylyeva, M.N, Svetlov, D, Svetlov, V, Perederina, A, Igarashi, N, Matsugaki, N, Wakatsuki, S, Tahirov, T.H, Vassylyev, D.G, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-07-01 | | Release date: | 2005-09-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Allosteric modulation of the RNA polymerase catalytic reaction is an essential component of transcription control by rifamycins.
Cell(Cambridge,Mass.), 122, 2005
|
|
4MQ9
 
 | | Crystal structure of Thermus thermophilus RNA polymerase holoenzyme in complex with GE23077 | | Descriptor: | (2Z)-2-methylbut-2-enoic acid, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Ho, M.X, Arnold, E, Ebright, R.H, Zhang, Y, Tuske, S. | | Deposit date: | 2013-09-16 | | Release date: | 2014-05-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | GE23077 binds to the RNA polymerase 'i' and 'i+1' sites and prevents the binding of initiating nucleotides.
Elife, 3, 2014
|
|
7MKI
 
 | | Cryo-EM structure of Escherichia coli RNA polymerase bound to lambda PR (-5G to C) promoter DNA | | Descriptor: | CHAPSO, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Saecker, R.M, Darst, S.A, Chen, J. | | Deposit date: | 2021-04-23 | | Release date: | 2021-09-29 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural origins of Escherichia coli RNA polymerase open promoter complex stability.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
4OIR
 
 | | Crystal structure of Thermus thermophilus RNA polymerase transcription initiation complex soaked with GE23077 and rifamycin SV | | Descriptor: | (2Z)-2-methylbut-2-enoic acid, 5'-D(*CP*CP*T*GP*CP*AP*TP*CP*CP*GP*TP*GP*AP*GP*TP*CP*GP*AP*GP*GP*G)-3', 5'-D(*TP*AP*TP*AP*AP*TP*GP*GP*GP*AP*GP*CP*TP*GP*TP*CP*AP*CP*GP*GP*AP*TP*GP*CP*AP*GP*G)-3', ... | | Authors: | Zhang, Y, Ebright, R.H, Arnold, E. | | Deposit date: | 2014-01-20 | | Release date: | 2014-05-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.105 Å) | | Cite: | GE23077 binds to the RNA polymerase 'i' and 'i+1' sites and prevents the binding of initiating nucleotides.
Elife, 3, 2014
|
|
7MKJ
 
 | | Cryo-EM structure of Escherichia coli RNA polymerase bound to T7A1 promoter DNA | | Descriptor: | CHAPSO, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Saecker, R.M, Darst, S.A, Chen, J. | | Deposit date: | 2021-04-23 | | Release date: | 2021-09-29 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural origins of Escherichia coli RNA polymerase open promoter complex stability.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7MKD
 
 | | Cryo-EM structure of Escherichia coli RNA polymerase bound to lambda PR promoter DNA (class 1) | | Descriptor: | CHAPSO, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Saecker, R.M, Darst, S.A, Chen, J. | | Deposit date: | 2021-04-23 | | Release date: | 2021-09-29 | | Last modified: | 2021-10-13 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural origins of Escherichia coli RNA polymerase open promoter complex stability.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7MKE
 
 | | Cryo-EM structure of Escherichia coli RNA polymerase bound to lambda PR promoter DNA (class 2) | | Descriptor: | CHAPSO, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Saecker, R.M, Darst, S.A, Chen, J. | | Deposit date: | 2021-04-23 | | Release date: | 2021-09-29 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural origins of Escherichia coli RNA polymerase open promoter complex stability.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
2C35
 
 | | Subunits Rpb4 and Rpb7 of human RNA polymerase II | | Descriptor: | DNA-DIRECTED RNA POLYMERASE II 16 KDA POLYPEPTIDE, DNA-DIRECTED RNA POLYMERASE II 19 KDA POLYPEPTIDE | | Authors: | Meka, H, Werner, F, Cordell, S.C, Onesti, S, Brick, P. | | Deposit date: | 2005-10-04 | | Release date: | 2005-11-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structure and RNA Binding of the Rpb4/Rpb7 Subunits of Human RNA Polymerase II.
Nucleic Acids Res., 33, 2005
|
|
1YV2
 
 | | Hepatitis C virus NS5B RNA-dependent RNA Polymerase genotype 2a | | Descriptor: | GLYCEROL, RNA dependent RNA polymerase, SULFATE ION | | Authors: | Biswal, B.K, Cherney, M.M, Wang, M, Chan, L, Yannopoulos, C.G, Bilimoria, D, Nicolas, O, Bedard, J, James, M.N.G. | | Deposit date: | 2005-02-14 | | Release date: | 2005-03-22 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structures of the RNA-dependent RNA Polymerase Genotype 2a of Hepatitis C Virus Reveal Two Conformations and Suggest Mechanisms of Inhibition by Non-nucleoside Inhibitors
J.Biol.Chem., 280, 2005
|
|
3HKZ
 
 | | The X-ray crystal structure of RNA polymerase from Archaea | | Descriptor: | DNA-directed RNA polymerase subunit 13, DNA-directed RNA polymerase subunit A', DNA-directed RNA polymerase subunit A'', ... | | Authors: | Murakami, K.S. | | Deposit date: | 2009-05-26 | | Release date: | 2009-06-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | The X-ray crystal structure of RNA polymerase from Archaea.
Nature, 451, 2008
|
|
