7NPI
 
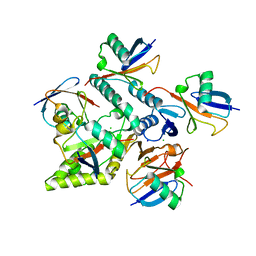 | | Crystal structure of Mindy2 (C266A) in complex with Lys48-linked penta-ubiquitin (K48-Ub5) | | Descriptor: | CHLORIDE ION, Polyubiquitin-C, SODIUM ION, ... | | Authors: | Lange, S.M, Armstrong, L.A, Kulathu, Y. | | Deposit date: | 2021-02-26 | | Release date: | 2021-09-15 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Mechanism of activation and regulation of deubiquitinase activity in MINDY1 and MINDY2.
Mol.Cell, 81, 2021
|
|
6JH1
 
 | |
6J62
 
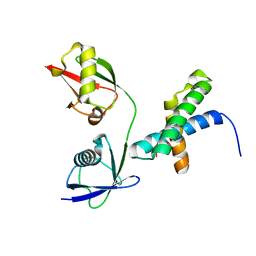 | | Crystal structure of mISG15/NS1B complex | | Descriptor: | Non-structural protein 1, Ubiquitin-like protein ISG15 | | Authors: | Jiang, Y.N, Wang, X.Q. | | Deposit date: | 2019-01-12 | | Release date: | 2019-12-25 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.486 Å) | | Cite: | Crystal structure of mISG15/NS1B complex
To Be Published
|
|
7OWD
 
 | |
7OWC
 
 | |
6JWI
 
 | | Yeast Npl4 in complex with Lys48-linked diubiquitin | | Descriptor: | BICINE, Nuclear protein localization protein 4, Ubiqutin, ... | | Authors: | Sato, Y, Fukai, S. | | Deposit date: | 2019-04-20 | | Release date: | 2019-12-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural insights into ubiquitin recognition and Ufd1 interaction of Npl4.
Nat Commun, 10, 2019
|
|
3PRM
 
 | | Structural analysis of a viral OTU domain protease from the Crimean-Congo Hemorrhagic Fever virus in complex with human ubiquitin | | Descriptor: | Polyubiquitin-B (Fragment), RNA-directed RNA polymerase L | | Authors: | Capodagli, G.C, McKercher, M.A, Baker, E.A, Masters, E.M, Brunzelle, J.S, Pegan, S.D. | | Deposit date: | 2010-11-30 | | Release date: | 2011-01-26 | | Last modified: | 2023-02-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural analysis of a viral ovarian tumor domain protease from the crimean-congo hemorrhagic Fever virus in complex with covalently bonded ubiquitin.
J.Virol., 85, 2011
|
|
7OJE
 
 | | Crystal structure of the covalent complex between Tribolium castaneum deubiquitinase ZUP and Ubiquitin-PA | | Descriptor: | 1,2-ETHANEDIOL, CITRIC ACID, Lys-63-specific deubiquitinase ZUFSP, ... | | Authors: | Pichlo, C, Hermanns, T, Hofmann, K, Baumann, U. | | Deposit date: | 2021-05-14 | | Release date: | 2022-02-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | A structural basis for the diverse linkage specificities within the ZUFSP deubiquitinase family.
Nat Commun, 13, 2022
|
|
3Q3F
 
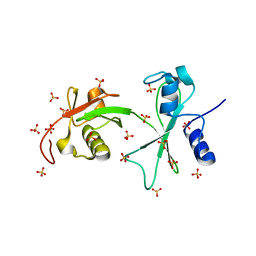 | | Engineering Domain-Swapped Binding Interfaces by Mutually Exclusive Folding: Insertion of Ubiquitin into position 103 of Barnase | | Descriptor: | Ribonuclease/Ubiquitin chimeric protein, SULFATE ION | | Authors: | Ha, J.-H, Karchin, J.M, Walker-Kopp, N, Huang, L.-S, Berry, E.A, Loh, S.N. | | Deposit date: | 2010-12-21 | | Release date: | 2012-01-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.169 Å) | | Cite: | Engineering domain-swapped binding interfaces by mutually exclusive folding.
J.Mol.Biol., 416, 2012
|
|
3NHE
 
 | |
3PSE
 
 | | Structure of a viral OTU domain protease bound to interferon-stimulated gene 15 (ISG15) | | Descriptor: | 1.7.6 3-bromanylpropan-1-amine, GLYCEROL, RNA polymerase, ... | | Authors: | Bacik, J.P, James, T.W, Frias-Staheli, N, Garcia-Sastre, A, Mark, B.L. | | Deposit date: | 2010-12-01 | | Release date: | 2011-01-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for the removal of ubiquitin and interferon-stimulated gene 15 by a viral ovarian tumor domain-containing protease.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3PT2
 
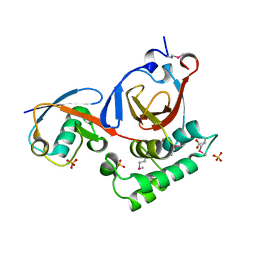 | | Structure of a viral OTU domain protease bound to Ubiquitin | | Descriptor: | 1.7.6 3-bromanylpropan-1-amine, ACETATE ION, RNA polymerase, ... | | Authors: | James, T.W, Bacik, J.P, Frias-Staheli, N, Garcia-Sastre, A, Mark, B.L. | | Deposit date: | 2010-12-02 | | Release date: | 2011-01-19 | | Last modified: | 2023-05-31 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for the removal of ubiquitin and interferon-stimulated gene 15 by a viral ovarian tumor domain-containing protease.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
7P7W
 
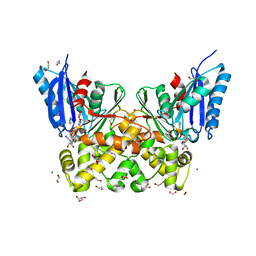 | | N-acetylglucosamine kinase from Plesiomonas shigelloides compexed with alpha-N-acetylglucosamine and ADP | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-alpha-D-glucopyranose, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Roy, S, Isupov, M.N, Harmer, N.J, Ames, J.R. | | Deposit date: | 2021-07-20 | | Release date: | 2022-08-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Spinning sugars in antigen biosynthesis: characterization of the Coxiella burnetii and Streptomyces griseus TDP-sugar epimerases
J.Biol.Chem., 2022
|
|
7P7I
 
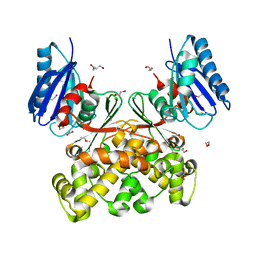 | | Native structure of N-acetylglucosamine kinase from Plesiomonas shigelloides | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, ... | | Authors: | Roy, S, Isupov, M.N, Harmer, N.J, Ames, J.R. | | Deposit date: | 2021-07-19 | | Release date: | 2022-07-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Spinning sugars in antigen biosynthesis: characterization of the Coxiella burnetii and Streptomyces griseus TDP-sugar epimerases
J.Biol.Chem., 2022
|
|
7P9L
 
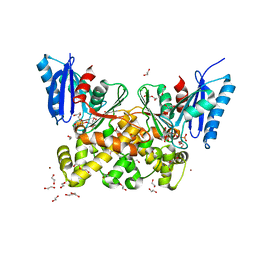 | | N-acetylglucosamine kinase from Plesiomonas shigelloides compexed with alpha-N-acetylglucosamine-6-phosphate | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-6-O-phosphono-beta-D-glucopyranose, ... | | Authors: | Roy, S, Isupov, M.N, Harmer, N.J, Ames, J.R. | | Deposit date: | 2021-07-27 | | Release date: | 2022-08-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Spinning sugars in antigen biosynthesis: characterization of the Coxiella burnetii and Streptomyces griseus TDP-sugar epimerases
J.Biol.Chem., 2022
|
|
7P9P
 
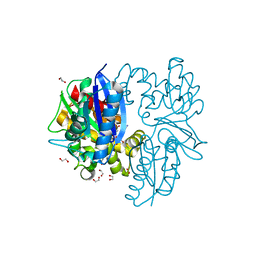 | | N-acetylglucosamine kinase from Plesiomonas shigelloides compexed with alpha-N-acetylglucosamine and AMP-PNP inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-alpha-D-glucopyranose, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Roy, S, Isupov, M.N, Harmer, N.J, Ames, J.R. | | Deposit date: | 2021-07-27 | | Release date: | 2022-08-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Spinning sugars in antigen biosynthesis: characterization of the Coxiella burnetii and Streptomyces griseus TDP-sugar epimerases
J.Biol.Chem., 2022
|
|
3PHW
 
 | | OTU Domain of Crimean Congo Hemorrhagic Fever Virus in complex with Ubiquitin | | Descriptor: | ETHANAMINE, RNA-directed RNA polymerase L, Ubiquitin-40S ribosomal protein S27a | | Authors: | Akutsu, M, Ye, Y, Virdee, S, Komander, D. | | Deposit date: | 2010-11-04 | | Release date: | 2011-02-02 | | Last modified: | 2014-08-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular basis for ubiquitin and ISG15 cross-reactivity in viral ovarian tumor domains.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
7PA1
 
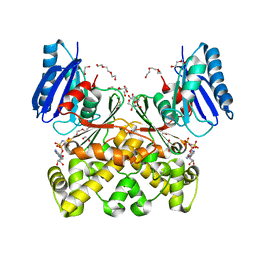 | | Structure of N-acetylglucosamine kinase from Plesiomonas shigelloides in complex with AMP-PNP in the absence of N-acetylglucoseamine substrate | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Roy, S, Isupov, M.N, Harmer, N.J, Ames, J.R. | | Deposit date: | 2021-07-28 | | Release date: | 2022-08-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Spinning sugars in antigen biosynthesis: characterization of the Coxiella burnetii and Streptomyces griseus TDP-sugar epimerases
J.Biol.Chem., 2022
|
|
7P9Y
 
 | | N-acetylglucosamine kinase from Plesiomonas shigelloides compexed with alpha-N-acetylglucosamine | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-alpha-D-glucopyranose, ... | | Authors: | Roy, S, Isupov, M.N, Harmer, N.J, Ames, J.R. | | Deposit date: | 2021-07-28 | | Release date: | 2022-08-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Spinning sugars in antigen biosynthesis: characterization of the Coxiella burnetii and Streptomyces griseus TDP-sugar epimerases
J.Biol.Chem., 2022
|
|
3OLM
 
 | | Structure and Function of a Ubiquitin Binding Site within the Catalytic Domain of a HECT Ubiquitin Ligase | | Descriptor: | E3 ubiquitin-protein ligase RSP5, Ubiquitin | | Authors: | Kim, H.C, Steffen, A, Chen, J, Huibregtse, J.M. | | Deposit date: | 2010-08-26 | | Release date: | 2011-03-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.495 Å) | | Cite: | Structure and function of a HECT domain ubiquitin-binding site.
Embo Rep., 12, 2011
|
|
7PI4
 
 | | FAK Protac GSK215 in complex with FAK and pVHL:ElonginC:ElonginB | | Descriptor: | (2S,4R)-4-hydroxy-1-((S)-2-(2-(4-(3-methoxy-4-((4-((2-(methylcarbamoyl)phenyl)amino)-5-(trifluoromethyl)pyridin-2-yl)amino)phenyl)piperazin-1-yl)acetamido)-3,3-dimethylbutanoyl)-N-((S)-1-(4-(4-methylthiazol-5-yl)phenyl)ethyl)pyrrolidine-2-carboxamide, 1,2-ETHANEDIOL, CALCIUM ION, ... | | Authors: | Chung, C. | | Deposit date: | 2021-08-19 | | Release date: | 2021-09-29 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Discovery and Characterisation of Highly Cooperative FAK-Degrading PROTACs.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
7PLO
 
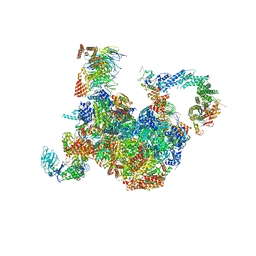 | | H. sapiens replisome-CUL2/LRR1 complex | | Descriptor: | Cell division control protein 45 homolog, Claspin, Cullin-2, ... | | Authors: | Jones, M.J, Yeeles, J.T.P, Deegan, T.D, Jenkyn-Bedford, M. | | Deposit date: | 2021-09-01 | | Release date: | 2021-11-10 | | Last modified: | 2023-10-18 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | A conserved mechanism for regulating replisome disassembly in eukaryotes.
Nature, 600, 2021
|
|
3MTN
 
 | | Usp21 in complex with a ubiquitin-based, USP21-specific inhibitor | | Descriptor: | CHLORIDE ION, GLYCEROL, UBIQUITIN VARIANT UBV.21.4, ... | | Authors: | Walker, J.R, Avvakumov, G.V, Xue, S, Li, Y, Ernst, A, Sidhu, S, Weigelt, J, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-04-30 | | Release date: | 2010-06-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A strategy for modulation of enzymes in the ubiquitin system.
Science, 339, 2013
|
|
3N3K
 
 | | The catalytic domain of USP8 in complex with a USP8 specific inhibitor | | Descriptor: | Ubiquitin, Ubiquitin carboxyl-terminal hydrolase 8, ZINC ION | | Authors: | Walker, J.R, Avvakumov, G.V, Xue, S, Li, Y, Allali-Hassani, A, Lam, R, Ernst, A, Sidhu, S, Weigelt, J, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-05-20 | | Release date: | 2010-06-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A strategy for modulation of enzymes in the ubiquitin system.
Science, 339, 2013
|
|
3PLV
 
 | | Structure of Hub-1 protein in complex with Snu66 peptide (HINDII) | | Descriptor: | 66 kDa U4/U6.U5 small nuclear ribonucleoprotein component, Ubiquitin-like modifier HUB1 | | Authors: | Mishra, S.K, Ammon, T, Popowicz, G.M, Krajewski, M, Nagel, R.J, Ares, M, Holak, T.A, Jentsch, S. | | Deposit date: | 2010-11-15 | | Release date: | 2011-06-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Role of the ubiquitin-like protein Hub1 in splice-site usage and alternative splicing.
Nature, 474, 2011
|
|
