4YF1
 
 | | 1.85 angstrom crystal structure of lmo0812 from Listeria monocytogenes EGD-e | | Descriptor: | CITRATE ANION, Lmo0812 protein, SODIUM ION | | Authors: | Krishna, S.N, Light, S.H, Filippova, E.V, Minasov, G, Kiryukhina, O, Jedrzejczak, R, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2015-02-24 | | Release date: | 2015-03-04 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | 1.85 angstrom crystal structure of lmo0812 from Listeria monocytogenes EGD-e
To Be Published
|
|
4FXI
 
 | |
4GQO
 
 | | 2.1 Angstrom resolution crystal structure of uncharacterized protein lmo0859 from Listeria monocytogenes EGD-e | | Descriptor: | Lmo0859 protein, TRIETHYLENE GLYCOL | | Authors: | Halavaty, A.S, Filippova, E.V, Minasov, G, Dubrovska, I, Winsor, J, Shuvalova, L, Peterson, S.N, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-08-23 | | Release date: | 2012-09-19 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | 2.1 Angstrom resolution crystal structure of uncharacterized protein lmo0859 from Listeria monocytogenes EGD-e
To be Published
|
|
5TLL
 
 | | Crystal Structure of the ER-alpha Ligand-binding Domain (Y537S) in Complex with (E)-2-chloro-4'-hydroxy-4-((hydroxyiminio)methyl)-[1,1'-biphenyl]-3-olate | | Descriptor: | 2-chloro-4-[(E)-(hydroxyimino)methyl][1,1'-biphenyl]-3,4'-diol, Estrogen receptor, NUCLEAR RECEPTOR COACTIVATOR 2 | | Authors: | Nwachukwu, J.C, Srinivasan, S, Bruno, N.E, Nowak, J, Kojetin, D.J, Elemento, O, Katzenellenbogen, J.A, Nettles, K.W. | | Deposit date: | 2016-10-11 | | Release date: | 2017-01-18 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.423 Å) | | Cite: | Systems Structural Biology Analysis of Ligand Effects on ER alpha Predicts Cellular Response to Environmental Estrogens and Anti-hormone Therapies.
Cell Chem Biol, 24, 2017
|
|
7Y81
 
 | | CryoEM structure of type III-E CRISPR Craspase gRAMP-crRNA complex bound to non-self RNA target | | Descriptor: | MAGNESIUM ION, Non-self RNA target, RAMP superfamily protein, ... | | Authors: | Zhang, J.T, Cui, N, Huang, H.D, Jia, N. | | Deposit date: | 2022-06-22 | | Release date: | 2022-12-14 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (2.54 Å) | | Cite: | Structural basis for the non-self RNA-activated protease activity of the type III-E CRISPR nuclease-protease Craspase.
Nat Commun, 13, 2022
|
|
7Y85
 
 | | CryoEM structure of type III-E CRISPR Craspase gRAMP-crRNA in complex with TPR-CHAT protease bound to self RNA target | | Descriptor: | CHAT domain protein, MAGNESIUM ION, RAMP superfamily protein, ... | | Authors: | Zhang, J.T, Cui, N, Huang, H.D, Jia, N. | | Deposit date: | 2022-06-22 | | Release date: | 2022-12-14 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (2.73 Å) | | Cite: | Structural basis for the non-self RNA-activated protease activity of the type III-E CRISPR nuclease-protease Craspase.
Nat Commun, 13, 2022
|
|
7Y82
 
 | | CryoEM structure of type III-E CRISPR Craspase gRAMP-crRNA complex bound to self RNA target | | Descriptor: | MAGNESIUM ION, RAMP superfamily protein, Self RNA target, ... | | Authors: | Zhang, J.T, Cui, N, Huang, H.D, Jia, N. | | Deposit date: | 2022-06-22 | | Release date: | 2022-12-14 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (2.83 Å) | | Cite: | Structural basis for the non-self RNA-activated protease activity of the type III-E CRISPR nuclease-protease Craspase.
Nat Commun, 13, 2022
|
|
7Y83
 
 | | CryoEM structure of type III-E CRISPR Craspase gRAMP-crRNA in complex with TPR-CHAT protease bound to non-self RNA target | | Descriptor: | CHAT domain protein, MAGNESIUM ION, RAMP superfamily protein, ... | | Authors: | Zhang, J.T, Cui, N, Huang, H.D, Jia, N. | | Deposit date: | 2022-06-22 | | Release date: | 2022-12-14 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (2.93 Å) | | Cite: | Structural basis for the non-self RNA-activated protease activity of the type III-E CRISPR nuclease-protease Craspase.
Nat Commun, 13, 2022
|
|
7Y80
 
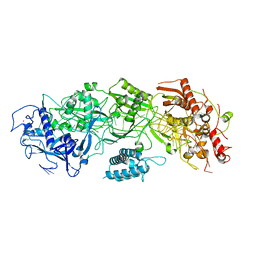 | | CryoEM structure of type III-E CRISPR Craspase gRAMP-crRNA binary complex | | Descriptor: | MAGNESIUM ION, RAMP superfamily protein, ZINC ION, ... | | Authors: | Zhang, J.T, Cui, N, Huang, H.D, Jia, N. | | Deposit date: | 2022-06-22 | | Release date: | 2022-12-14 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (2.71 Å) | | Cite: | Structural basis for the non-self RNA-activated protease activity of the type III-E CRISPR nuclease-protease Craspase.
Nat Commun, 13, 2022
|
|
7Y84
 
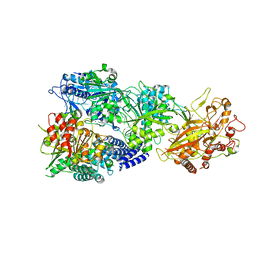 | | CryoEM structure of type III-E CRISPR Craspase gRAMP-crRNA in complex with TPR-CHAT protease | | Descriptor: | CHAT domain protein, MAGNESIUM ION, RAMP superfamily protein, ... | | Authors: | Zhang, J.T, Cui, N, Huang, H.D, Jia, N. | | Deposit date: | 2022-06-22 | | Release date: | 2022-12-14 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (2.61 Å) | | Cite: | Structural basis for the non-self RNA-activated protease activity of the type III-E CRISPR nuclease-protease Craspase.
Nat Commun, 13, 2022
|
|
6CVK
 
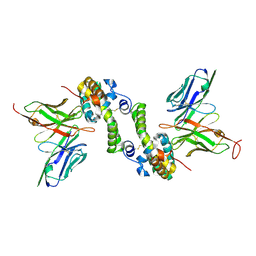 | |
4JXF
 
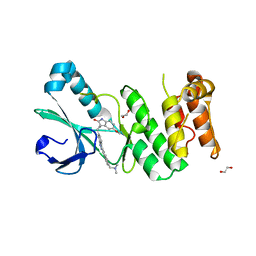 | | Crystal Structure of PLK4 Kinase with an inhibitor: 400631 ((1R,2S)-2-{3-[(E)-2-{4-[(DIMETHYLAMINO)METHYL]PHENYL}ETHENYL]-2H-INDAZOL-6-YL}-5'-METHOXYSPIRO[CYCLOPROPANE-1,3'-INDOL]-2'(1'H)-ONE) | | Descriptor: | (1R,2S)-2-{3-[(E)-2-{4-[(dimethylamino)methyl]phenyl}ethenyl]-2H-indazol-6-yl}-5'-methoxyspiro[cyclopropane-1,3'-indol]-2'(1'H)-one, 1,2-ETHANEDIOL, Serine/threonine-protein kinase PLK4 | | Authors: | Qiu, W, Plotnikova, O, Feher, M, Awrey, D.E, Chirgadze, N.Y. | | Deposit date: | 2013-03-28 | | Release date: | 2014-04-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of PLK4 Kinase with an inhibitor: 400631
To be Published
|
|
4JYI
 
 | | Crystal structure of RARbeta LBD in complex with selective partial agonist BMS641 [3-chloro-4-[(E)-2-(5,5-dimethyl-8-phenyl-5,6-dihydronaphthalen-2-yl)ethenyl]benzoic acid] | | Descriptor: | 3-chloro-4-[(E)-2-(5,5-dimethyl-8-phenyl-5,6-dihydronaphthalen-2-yl)ethenyl]benzoic acid, CITRATE ANION, Nuclear receptor coactivator 1, ... | | Authors: | Nadendla, E.K, Teyssier, C, Germain, P, Delfosse, V, Bourguet, W. | | Deposit date: | 2013-03-29 | | Release date: | 2014-03-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | An Unexpected Mode Of Binding Defines BMS948 as A Full Retinoic Acid Receptor beta (RAR beta , NR1B2) Selective Agonist.
Plos One, 10, 2015
|
|
2KU0
 
 | | Inhibitor Induced Structural Change in the HCV IRES Domain IIa RNA | | Descriptor: | (7S)-7-[(dimethylamino)methyl]-1-[3-(dimethylamino)propyl]-7,8-dihydro-1H-furo[3,2-e]benzimidazol-2-amine, HCV IRES Domain IIa RNA | | Authors: | Paulsen, R.B, Seth, P.P, Swayze, E.E, Griffey, R.H, Skalicky, J.J, Cheatham III, T.E, Davis, D.R. | | Deposit date: | 2010-02-10 | | Release date: | 2010-04-28 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Inhibitor-induced structural change in the HCV IRES domain IIa RNA.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
1EEO
 
 | | CRYSTAL STRUCTURE OF PROTEIN TYROSINE PHOSPHATASE 1B COMPLEXED WITH ACETYL-E-L-E-F-PTYR-M-D-Y-E-NH2 | | Descriptor: | ACETYL-E-L-E-F-PTYR-M-D-Y-E-NH2 PEPTIDE, MAGNESIUM ION, PROTEIN TYROSINE PHOSPHATASE 1B | | Authors: | Sarmiento, M, Puius, Y.A, Vetter, S.W, Lawrence, D.S, Almo, S.C, Zhang, Z.Y. | | Deposit date: | 2000-02-01 | | Release date: | 2001-02-01 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis of plasticity in protein tyrosine phosphatase 1B substrate recognition.
Biochemistry, 39, 2000
|
|
5XMM
 
 | | FLA-E*01801-167W/S | | Descriptor: | Beta-2-microglobulin, Gag polyprotein, MHC class I antigen alpha chain | | Authors: | Liang, R, Sun, Y, Wang, J, Wu, Y, Zhang, N, Xia, C. | | Deposit date: | 2017-05-15 | | Release date: | 2017-12-13 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Major Histocompatibility Complex Class I (FLA-E*01801) Molecular Structure in Domestic Cats Demonstrates Species-Specific Characteristics in Presenting Viral Antigen Peptides
J. Virol., 92, 2018
|
|
2L61
 
 | | Protein and metal cluster structure of the wheat metallothionein domain g-Ec-1. The second part of the puzzle. | | Descriptor: | CADMIUM ION, EC protein I/II | | Authors: | Loebus, J, Peroza, E.A, Bluethgen, N, Fox, T, Meyer-Klaucke, W, Zerbe, O, Freisinger, E. | | Deposit date: | 2010-11-12 | | Release date: | 2011-05-25 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Protein and metal cluster structure of the wheat metallothionein domain gamma-E(c)-1: the second part of the puzzle.
J.Biol.Inorg.Chem., 16, 2011
|
|
4HJS
 
 | |
4HN4
 
 | | Tryptophan synthase in complex with alpha aminoacrylate E(A-A) form and the F9 inhibitor in the alpha site | | Descriptor: | 2-({[4-(TRIFLUOROMETHOXY)PHENYL]SULFONYL}AMINO)ETHYL DIHYDROGEN PHOSPHATE, 2-{[(E)-{3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene]amino}prop-2-enoic acid, BICINE, ... | | Authors: | Hilario, E, Niks, D, Dunn, M.F, Mueller, L.J, Fan, L. | | Deposit date: | 2012-10-18 | | Release date: | 2013-12-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Allostery and substrate channeling in the tryptophan synthase bienzyme complex: evidence for two subunit conformations and four quaternary states.
Biochemistry, 52, 2013
|
|
4HJT
 
 | |
4HPX
 
 | | Crystal structure of Tryptophan Synthase at 1.65 A resolution in complex with alpha aminoacrylate E(A-A) and benzimidazole in the beta site and the F9 inhibitor in the alpha site | | Descriptor: | 2-({[4-(TRIFLUOROMETHOXY)PHENYL]SULFONYL}AMINO)ETHYL DIHYDROGEN PHOSPHATE, 2-{[(E)-{3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene]amino}prop-2-enoic acid, BENZIMIDAZOLE, ... | | Authors: | Hilario, E, Niks, D, Dunn, M.F, Mueller, L.J, Fan, L. | | Deposit date: | 2012-10-24 | | Release date: | 2013-12-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Allostery and substrate channeling in the tryptophan synthase bienzyme complex: evidence for two subunit conformations and four quaternary states.
Biochemistry, 52, 2013
|
|
8YRJ
 
 | | Mouse Fc epsilon RI | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, High affinity immunoglobulin epsilon receptor subunit alpha, High affinity immunoglobulin epsilon receptor subunit beta, ... | | Authors: | Zhang, Z, Yui, M, Ohto, U, Shimizu, T. | | Deposit date: | 2024-03-21 | | Release date: | 2024-07-17 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.87 Å) | | Cite: | Architecture of the high-affinity immunoglobulin E receptor.
Sci.Signal., 17, 2024
|
|
5FMG
 
 | | Structure and function based design of Plasmodium-selective proteasome inhibitors | | Descriptor: | (2S)-N-[(E,2S)-1-(1H-indol-3-yl)-4-methylsulfonyl-but-3-en-2-yl]-2-[[(2S)-3-(1H-indol-3-yl)-2-(2-morpholin-4-ylethanoylamino)propanoyl]amino]-4-methyl-pentanamide, BETA3 PROTEASOME SUBUNIT, PUTATIVE, ... | | Authors: | Li, H, O'Donoghue, A.J, van der Linden, W.A, Xie, S.C, Yoo, E, Foe, I.T, Tilley, L, Craik, C.S, da Fonseca, P.C.A, Bogyo, M. | | Deposit date: | 2015-11-04 | | Release date: | 2016-03-02 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structure and Function Based Design of Plasmodium-Selective Proteasome Inhibitors
Nature, 530, 2016
|
|
3JCX
 
 | | Canine Parvovirus complexed with Fab E | | Descriptor: | Capsid protein 2, Fab E heavy chain, Fab E light chain | | Authors: | Organtini, L.J, Iketani, S, Huang, K, Ashley, R.E, Makhov, A.M, Conway, J.F, Parrish, C.R, Hafenstein, S. | | Deposit date: | 2016-03-21 | | Release date: | 2016-07-20 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Near-Atomic Resolution Structure of a Highly Neutralizing Fab Bound to Canine Parvovirus.
J.Virol., 90, 2016
|
|
2L62
 
 | | Protein and metal cluster structure of the wheat metallothionein domain g-Ec-1. The second part of the puzzle. | | Descriptor: | EC protein I/II, ZINC ION | | Authors: | Loebus, J, Peroza, E.A, Bluethgen, N, Fox, T, Meyer-Klaucke, W, Zerbe, O, Freisinger, E. | | Deposit date: | 2010-11-12 | | Release date: | 2011-05-25 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Protein and metal cluster structure of the wheat metallothionein domain gamma-E(c)-1: the second part of the puzzle.
J.Biol.Inorg.Chem., 16, 2011
|
|
