3G4C
 
 | |
5GKK
 
 | |
6I0K
 
 | | Structure of quinolinate synthase in complex with 4-mercaptophthalic acid | | Descriptor: | 4-mercaptoidenecyclohexa-2,5-diene-1,2-dicarboxylic acid, CHLORIDE ION, IRON/SULFUR CLUSTER, ... | | Authors: | Volbeda, A, Fontecilla-Camps, J.C. | | Deposit date: | 2018-10-26 | | Release date: | 2019-03-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Design of specific inhibitors of quinolinate synthase based on [4Fe-4S] cluster coordination.
Chem.Commun.(Camb.), 55, 2019
|
|
1R2W
 
 | | Coordinates of L11 with 58nts of 23S rRNA fitted into the cryo-EM map of the 70S ribosome | | Descriptor: | 50S ribosomal protein L11, 58nts of 23S rRNA | | Authors: | Valle, M, Zavialov, A, Li, W, Stagg, S.M, Sengupta, J, Nielsen, R.C, Nissen, P, Harvey, S.C, Ehrenberg, M, Frank, J. | | Deposit date: | 2003-09-30 | | Release date: | 2003-11-04 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (9 Å) | | Cite: | Incorporation of aminoacyl-tRNA into the ribosome as seen by cryo-electron Microscopy
Nat.Struct.Biol., 10, 2003
|
|
3CNN
 
 | | GTP-bound structure of TM YlqF | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, Putative uncharacterized protein | | Authors: | Kim, D.J, Jang, J.Y, Yoon, H.-J, Suh, S.W. | | Deposit date: | 2008-03-26 | | Release date: | 2008-06-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of YlqF, a circularly permuted GTPase: Implications for its GTPase activation in 50 S ribosomal subunit assembly
Proteins, 72, 2008
|
|
1T7L
 
 | |
2H4F
 
 | | Sir2-p53 peptide-NAD+ | | Descriptor: | Cellular tumor antigen p53, NAD-dependent deacetylase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Hoff, K.G, Avalos, J.L, Sens, K, Wolberger, C. | | Deposit date: | 2006-05-24 | | Release date: | 2006-09-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Insights into the Sirtuin Mechanism from Ternary Complexes Containing NAD(+) and Acetylated Peptide.
Structure, 14, 2006
|
|
5FF3
 
 | | HydE from T. maritima in complex with 4R-TCA | | Descriptor: | 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, CHLORIDE ION, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Rohac, R, Amara, P, Benjdia, A, Martin, L, Ruffie, P, Favier, A, Berteau, O, Mouesca, J.M, Fontecilla-Camps, J.C, Nicolet, Y. | | Deposit date: | 2015-12-17 | | Release date: | 2016-04-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | Carbon-sulfur bond-forming reaction catalysed by the radical SAM enzyme HydE.
Nat.Chem., 8, 2016
|
|
2H4H
 
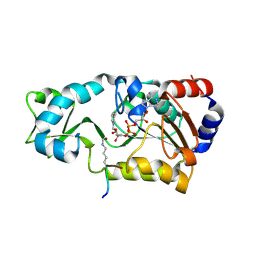 | | Sir2 H116Y mutant-p53 peptide-NAD | | Descriptor: | Cellular tumor antigen p53, NAD-dependent deacetylase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Hoff, K.G, Avalos, J.L, Sens, K, Wolberger, C. | | Deposit date: | 2006-05-24 | | Release date: | 2006-09-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Insights into the Sirtuin Mechanism from Ternary Complexes Containing NAD(+) and Acetylated Peptide.
Structure, 14, 2006
|
|
1VHO
 
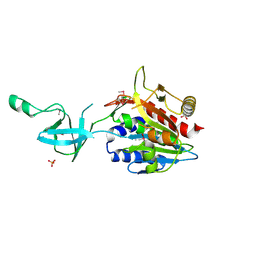 | |
1VHF
 
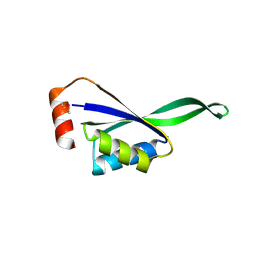 | |
3H3E
 
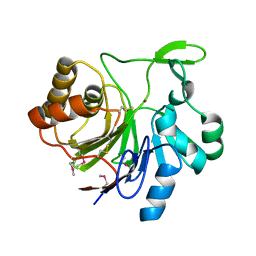 | | Crystal structure of Tm1679, A METAL-DEPENDENT HYDROLASE OF THE BETA-LACTAMASE SUPERFAMILY | | Descriptor: | ZINC ION, uncharacterized protein Tm1679 | | Authors: | Cooper, D.R, Olekhnovitch, N, Derewenda, U, Derewenda, Z.S, Integrated Center for Structure and Function Innovation (ISFI) | | Deposit date: | 2009-04-16 | | Release date: | 2009-07-14 | | Last modified: | 2021-10-13 | | Method: | X-RAY DIFFRACTION (2.749 Å) | | Cite: | Crystal structure of Tm1679, A METAL-DEPENDENT HYDROLASE OF THE BETA-LACTAMASE SUPERFAMILY
To be Published
|
|
2KA7
 
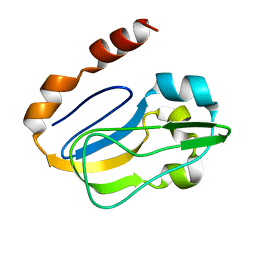 | | NMR solution structure of TM0212 at 40 C | | Descriptor: | Glycine cleavage system H protein | | Authors: | Pedrini, B, Herrmann, T, Mohanty, B, Geralt, M, Wilson, I, Wuthrich, K, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2008-10-31 | | Release date: | 2009-01-13 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | The J-UNIO protocol for automated protein structure determination by NMR in solution.
J.Biomol.Nmr, 53, 2012
|
|
2ZXB
 
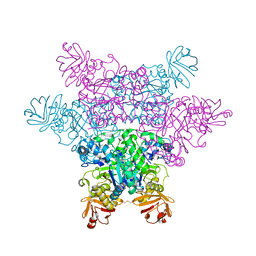 | | alpha-L-fucosidase complexed with inhibitor, ph-6FNJ | | Descriptor: | (2S,3R,4S,5R)-2-benzylpiperidine-3,4,5-triol, Alpha-L-fucosidase, putative | | Authors: | Wu, H.-J, Ko, T.-P, Ho, C.-W, Lin, C.-H, Wang, A.H.-J. | | Deposit date: | 2008-12-22 | | Release date: | 2009-12-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Structural basis of alpha-fucosidase inhibition by iminocyclitols with K(i) values in the micro- to picomolar range.
Angew.Chem.Int.Ed.Engl., 49, 2010
|
|
2ZXA
 
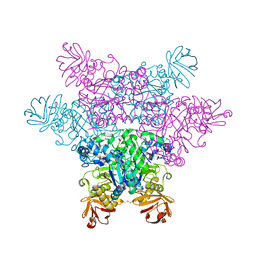 | | alpha-L-fucosidase complexed with inhibitor, FNJ-acetyl | | Descriptor: | Alpha-L-fucosidase, putative, N-{[(2R,3R,4R,5R,6S)-3,4,5-trihydroxy-6-methylpiperidin-2-yl]methyl}acetamide | | Authors: | Wu, H.-J, Ko, T.-P, Ho, C.-W, Lin, C.-H, Wang, A.H.-J. | | Deposit date: | 2008-12-22 | | Release date: | 2009-12-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Structural basis of alpha-fucosidase inhibition by iminocyclitols with K(i) values in the micro- to picomolar range.
Angew.Chem.Int.Ed.Engl., 49, 2010
|
|
2ZX5
 
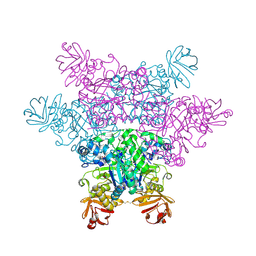 | | alpha-L-fucosidase complexed with inhibitor, F10 | | Descriptor: | 3-(1H-indol-3-yl)-N-{[(2R,3R,4R,5R,6S)-3,4,5-trihydroxy-6-methylpiperidin-2-yl]methyl}propanamide, Alpha-L-fucosidase, putative | | Authors: | Wu, H.-J, Ko, T.-P, Ho, C.-W, Lin, C.-H, Wang, A.H.-J. | | Deposit date: | 2008-12-19 | | Release date: | 2009-12-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural basis of alpha-fucosidase inhibition by iminocyclitols with K(i) values in the micro- to picomolar range.
Angew.Chem.Int.Ed.Engl., 49, 2010
|
|
2ZX7
 
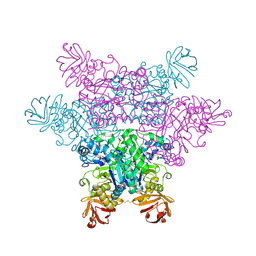 | | alpha-L-fucosidase complexed with inhibitor, F10-2C | | Descriptor: | Alpha-L-fucosidase, putative, N-{[(2R,3R,4R,5R,6S)-3,4,5-trihydroxy-6-methylpiperidin-2-yl]methyl}-1H-indole-2-carboxamide | | Authors: | Wu, H.-J, Ko, T.-P, Ho, C.-W, Lin, C.-H, Wang, A.H.-J. | | Deposit date: | 2008-12-19 | | Release date: | 2009-12-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Structural basis of alpha-fucosidase inhibition by iminocyclitols with K(i) values in the micro- to picomolar range.
Angew.Chem.Int.Ed.Engl., 49, 2010
|
|
2ZX8
 
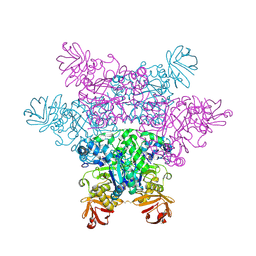 | | alpha-L-fucosidase complexed with inhibitor, F10-2C-O | | Descriptor: | Alpha-L-fucosidase, putative, N-{[(2R,3R,4R,5R,6S)-3,4,5-trihydroxy-6-methylpiperidin-2-yl]methyl}-1-benzofuran-2-carboxamide | | Authors: | Wu, H.-J, Ko, T.-P, Ho, C.-W, Lin, C.-H, Wang, A.H.-J. | | Deposit date: | 2008-12-19 | | Release date: | 2009-12-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Structural basis of alpha-fucosidase inhibition by iminocyclitols with K(i) values in the micro- to picomolar range.
Angew.Chem.Int.Ed.Engl., 49, 2010
|
|
2ZX9
 
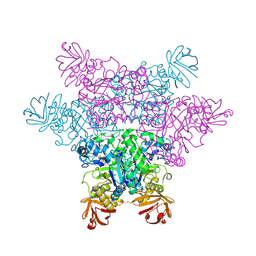 | | alpha-L-fucosidase complexed with inhibitor, B4 | | Descriptor: | (2S)-2-cyclopentyl-2-phenyl-N-{[(2R,3R,4R,5R,6S)-3,4,5-trihydroxy-6-methylpiperidin-2-yl]methyl}ethanamide, Alpha-L-fucosidase, putative | | Authors: | Wu, H.-J, Ko, T.-P, Ho, C.-W, Lin, C.-H, Wang, A.H.-J. | | Deposit date: | 2008-12-22 | | Release date: | 2009-12-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Structural basis of alpha-fucosidase inhibition by iminocyclitols with K(i) values in the micro- to picomolar range.
Angew.Chem.Int.Ed.Engl., 49, 2010
|
|
2ZX6
 
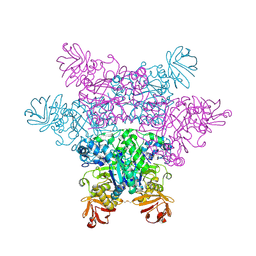 | | alpha-L-fucosidase complexed with inhibitor, F10-1C | | Descriptor: | 2-(1H-indol-3-yl)-N-{[(2R,3R,4R,5R,6S)-3,4,5-trihydroxy-6-methylpiperidin-2-yl]methyl}acetamide, Alpha-L-fucosidase, putative | | Authors: | Wu, H.-J, Ko, T.-P, Ho, C.-W, Lin, C.-H, Wang, A.H.-J. | | Deposit date: | 2008-12-19 | | Release date: | 2009-12-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Structural basis of alpha-fucosidase inhibition by iminocyclitols with K(i) values in the micro- to picomolar range.
Angew.Chem.Int.Ed.Engl., 49, 2010
|
|
1YC5
 
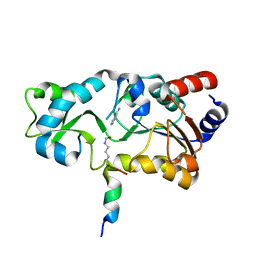 | | Sir2-p53 peptide-nicotinamide | | Descriptor: | Cellular tumor antigen p53 peptide, NAD-dependent deacetylase, NICOTINAMIDE, ... | | Authors: | Avalos, J.L, Bever, M.K, Wolberger, C. | | Deposit date: | 2004-12-21 | | Release date: | 2005-04-26 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Mechanism of sirtuin inhibition by nicotinamide: altering the NAD(+) cosubstrate specificity of a Sir2 enzyme.
Mol.Cell, 17, 2005
|
|
1W5F
 
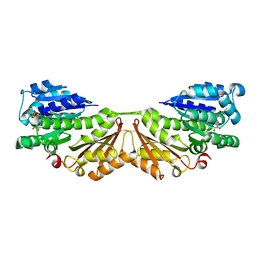 | | FtsZ, T7 mutated, domain swapped (T. maritima) | | Descriptor: | CELL DIVISION PROTEIN FTSZ, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER | | Authors: | Oliva, M.A, Cordell, S.C, Lowe, J. | | Deposit date: | 2004-08-06 | | Release date: | 2004-12-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Insights Into Ftsz Protofilament Formation
Nat.Struct.Mol.Biol., 11, 2004
|
|
3BQ5
 
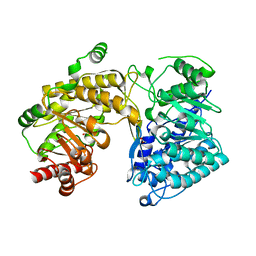 | | Crystal Structure of T. maritima Cobalamin-Independent Methionine Synthase complexed with Zn2+ and Homocysteine (Monoclinic) | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-4-MERCAPTO-BUTYRIC ACID, 5-methyltetrahydropteroyltriglutamate-homocysteine methyltransferase, ... | | Authors: | Pejchal, R, Smith, J.L, Ludwig, M.L. | | Deposit date: | 2007-12-19 | | Release date: | 2008-03-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Metal active site elasticity linked to activation of homocysteine in methionine synthases.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
1Z4A
 
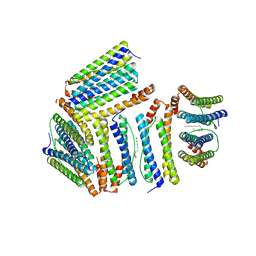 | |
3BQ6
 
 | |
