9JNZ
 
 | | Structure of isw1-nucleosome complex in Apo state | | Descriptor: | CHLORIDE ION, DNA (146-MER), Histone H2A, ... | | Authors: | Sia, Y, Pan, H, Chen, Z. | | Deposit date: | 2024-09-24 | | Release date: | 2025-04-09 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural insights into chromatin remodeling by ISWI during active ATP hydrolysis.
Science, 388, 2025
|
|
9JNW
 
 | | Structure of isw1-nucleosome complex in ADP+ state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA (147-MER), Histone H2A, ... | | Authors: | Sia, Y, Pan, H, Chen, Z. | | Deposit date: | 2024-09-24 | | Release date: | 2025-04-09 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural insights into chromatin remodeling by ISWI during active ATP hydrolysis.
Science, 388, 2025
|
|
9JNU
 
 | | Structure of isw1-nucleosome complex in ADP state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA (146-MER), Histone H2A, ... | | Authors: | Sia, Y, Pan, H, Chen, Z. | | Deposit date: | 2024-09-24 | | Release date: | 2025-04-09 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Structural insights into chromatin remodeling by ISWI during active ATP hydrolysis.
Science, 388, 2025
|
|
5MPB
 
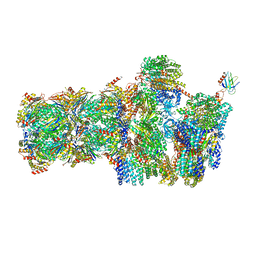 | | 26S proteasome in presence of AMP-PNP (s3) | | Descriptor: | 26S protease regulatory subunit 4 homolog, 26S protease regulatory subunit 6A, 26S protease regulatory subunit 6B homolog, ... | | Authors: | Wehmer, M, Rudack, T, Beck, F, Aufderheide, A, Pfeifer, G, Plitzko, J.M, Foerster, F, Schulten, K, Baumeister, W, Sakata, E. | | Deposit date: | 2016-12-16 | | Release date: | 2017-03-08 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (7.8 Å) | | Cite: | Structural insights into the functional cycle of the ATPase module of the 26S proteasome.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
8BGJ
 
 | |
7NMI
 
 | |
6TUN
 
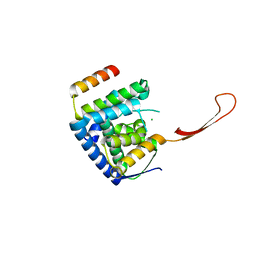 | | Helicase domain complex | | Descriptor: | CDK-activating kinase assembly factor MAT1, CHLORIDE ION, General transcription and DNA repair factor IIH helicase subunit XPD | | Authors: | Sauer, F, Kisker, C. | | Deposit date: | 2020-01-07 | | Release date: | 2020-11-11 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | In TFIIH the Arch domain of XPD is mechanistically essential for transcription and DNA repair.
Nat Commun, 11, 2020
|
|
7AO9
 
 | | Structure of the core MTA1/HDAC1/MBD2 NURD deacetylase complex | | Descriptor: | Histone deacetylase 1, INOSITOL HEXAKISPHOSPHATE, Metastasis-associated protein MTA1, ... | | Authors: | Millard, C.J, Fairall, L, Ragan, T.J, Savva, C.G, Schwabe, J.W.R. | | Deposit date: | 2020-10-14 | | Release date: | 2020-11-11 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (6.1 Å) | | Cite: | The topology of chromatin-binding domains in the NuRD deacetylase complex.
Nucleic Acids Res., 48, 2020
|
|
7AO8
 
 | | Structure of the MTA1/HDAC1/MBD2 NURD deacetylase complex | | Descriptor: | Histone deacetylase 1, INOSITOL HEXAKISPHOSPHATE, Metastasis-associated protein MTA1, ... | | Authors: | Millard, C.J, Fairall, L, Ragan, T.J, Savva, C.G, Schwabe, J.W.R. | | Deposit date: | 2020-10-14 | | Release date: | 2020-11-11 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | The topology of chromatin-binding domains in the NuRD deacetylase complex.
Nucleic Acids Res., 48, 2020
|
|
8OW1
 
 | | Cryo-EM structure of the yeast Inner kinetochore bound to a CENP-A nucleosome. | | Descriptor: | C0N3, Centromere DNA-binding protein complex CBF3 subunit B, Centromere DNA-binding protein complex CBF3 subunit C, ... | | Authors: | Dendooven, T.D, Zhang, Z, Yang, J, McLaughlin, S, Schwabb, J, Scheres, S, Yatskevich, S, Barford, D. | | Deposit date: | 2023-04-26 | | Release date: | 2023-08-09 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Cryo-EM structure of the complete inner kinetochore of the budding yeast point centromere.
Sci Adv, 9, 2023
|
|
9KBD
 
 | |
7ZBZ
 
 | |
7Z8V
 
 | | CAND1-SCF-SKP2 (SKP1deldel) CAND1 engaged SCF rocked | | Descriptor: | Cullin-1, Cullin-associated NEDD8-dissociated protein 1, E3 ubiquitin-protein ligase RBX1, ... | | Authors: | Baek, K, Schulman, B.A. | | Deposit date: | 2022-03-18 | | Release date: | 2023-04-19 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Systemwide disassembly and assembly of SCF ubiquitin ligase complexes.
Cell, 186, 2023
|
|
7Z8T
 
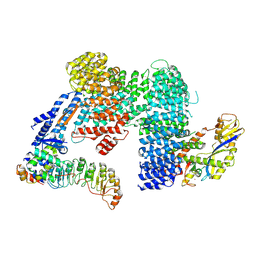 | | CAND1-SCF-SKP2 CAND1 engaged SCF rocked | | Descriptor: | Cullin-1, Cullin-associated NEDD8-dissociated protein 1, E3 ubiquitin-protein ligase RBX1, ... | | Authors: | Baek, K, Schulman, B.A. | | Deposit date: | 2022-03-18 | | Release date: | 2023-04-19 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Systemwide disassembly and assembly of SCF ubiquitin ligase complexes.
Cell, 186, 2023
|
|
7ZBW
 
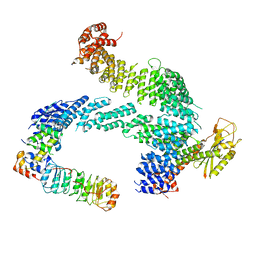 | | CAND1-SCF-SKP2 CAND1 rolling-2 SCF engaged | | Descriptor: | Cullin-1, Cullin-associated NEDD8-dissociated protein 1, E3 ubiquitin-protein ligase RBX1, ... | | Authors: | Baek, K, Schulman, B.A. | | Deposit date: | 2022-03-24 | | Release date: | 2023-04-19 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Systemwide disassembly and assembly of SCF ubiquitin ligase complexes.
Cell, 186, 2023
|
|
7K5Y
 
 | |
4WQO
 
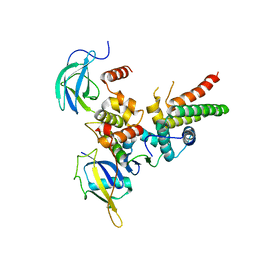 | | Structure of VHL-EloB-EloC-Cul2 | | Descriptor: | Cullin-2, Transcription elongation factor B polypeptide 1, Transcription elongation factor B polypeptide 2, ... | | Authors: | Nguyen, H.C, Xiong, Y. | | Deposit date: | 2014-10-22 | | Release date: | 2015-03-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Insights into Cullin-RING E3 Ubiquitin Ligase Recruitment: Structure of the VHL-EloBC-Cul2 Complex.
Structure, 23, 2015
|
|
3SMO
 
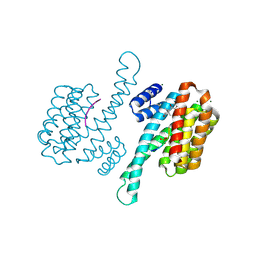 | | Crystal structure of human 14-3-3 sigma C38V/N166H in complex with TASK-3 peptide and stabilizer Fusicoccin J aglycone | | Descriptor: | 14-3-3 protein sigma, Fusicoccin J aglycone, MAGNESIUM ION, ... | | Authors: | Anders, C, Schumacher, B, Ottmann, C. | | Deposit date: | 2011-06-28 | | Release date: | 2012-07-11 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A semisynthetic fusicoccane stabilizes a protein-protein interaction and enhances the expression of K+ channels at the cell surface.
Chem. Biol., 20, 2013
|
|
3SML
 
 | | Crystal structure of human 14-3-3 sigma C38N/N166H in complex with TASK-3 peptide and stabilizer Fusicoccin A aglycone | | Descriptor: | 14-3-3 protein sigma, CHLORIDE ION, Fusicoccin A aglycone, ... | | Authors: | Anders, C, Schumacher, B, Ottmann, C. | | Deposit date: | 2011-06-28 | | Release date: | 2012-07-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A semisynthetic fusicoccane stabilizes a protein-protein interaction and enhances the expression of K+ channels at the cell surface.
Chem. Biol., 20, 2013
|
|
3DPL
 
 | |
6R7N
 
 | | Structural basis of Cullin-2 RING E3 ligase regulation by the COP9 signalosome | | Descriptor: | COP9 signalosome complex subunit 1, COP9 signalosome complex subunit 2, COP9 signalosome complex subunit 3, ... | | Authors: | Faull, S.V, Lau, A.M.C, Martens, C, Ahdash, Z, Yebenes, H, Schmidt, C, Beuron, F, Cronin, N.B, Morris, E.P, Politis, A. | | Deposit date: | 2019-03-29 | | Release date: | 2019-08-28 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (6.5 Å) | | Cite: | Structural basis of Cullin 2 RING E3 ligase regulation by the COP9 signalosome.
Nat Commun, 10, 2019
|
|
6R7I
 
 | | Structural basis of Cullin-2 RING E3 ligase regulation by the COP9 signalosome | | Descriptor: | COP9 signalosome complex subunit 1, COP9 signalosome complex subunit 2, COP9 signalosome complex subunit 3, ... | | Authors: | Faull, S.F, Lau, A.M.C, Beuron, F, Cronin, N.B, Morris, E.P, Politis, A. | | Deposit date: | 2019-03-28 | | Release date: | 2019-08-28 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (5.9 Å) | | Cite: | Structural basis of Cullin 2 RING E3 ligase regulation by the COP9 signalosome.
Nat Commun, 10, 2019
|
|
8IHN
 
 | | Cryo-EM structure of the Rpd3S core complex | | Descriptor: | CALCIUM ION, Chromatin modification-related protein EAF3, Histone H3, ... | | Authors: | Zhang, Y, Gang, C. | | Deposit date: | 2023-02-23 | | Release date: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.37 Å) | | Cite: | Structural basis for nucleosome binding and catalysis by the yeast Rpd3S/HDAC holoenzyme.
Cell Res., 33, 2023
|
|
8V6V
 
 | | Cryo-EM structure of doubly-bound SNF2h-nucleosome complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Histone H2A type 1, Histone H2B, ... | | Authors: | Chio, U.S, Palovcak, E, Armache, J.P, Narlikar, G.J, Cheng, Y. | | Deposit date: | 2023-12-03 | | Release date: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Functionalized graphene-oxide grids enable high-resolution cryo-EM structures of the SNF2h-nucleosome complex without crosslinking.
Nat Commun, 15, 2024
|
|
8V4Y
 
 | | Cryo-EM structure of singly-bound SNF2h-nucleosome complex with SNF2h at inactive SHL2 (conformation 1) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, Histone H2A type 1, ... | | Authors: | Chio, U.S, Palovcak, E, Armache, J.P, Narlikar, G.J, Cheng, Y. | | Deposit date: | 2023-11-29 | | Release date: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Functionalized graphene-oxide grids enable high-resolution cryo-EM structures of the SNF2h-nucleosome complex without crosslinking.
Nat Commun, 15, 2024
|
|
