6QR2
 
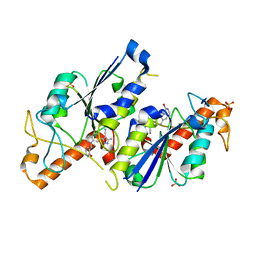 | | Crystal structure of TrmD, a tRNA-(N1G37) methyltransferase, from Mycobacterium abscessus in complex with inhibitor | | Descriptor: | 5-azanyl-3-[1-[(2-oxidanylpyridin-3-yl)methyl]indol-6-yl]-1~{H}-pyrazole-4-carbonitrile, SULFATE ION, tRNA (guanine-N(1)-)-methyltransferase | | Authors: | Thomas, S.E, Whitehouse, A.J, Coyne, A.G, Abell, C, Mendes, V, Blundell, T.L. | | Deposit date: | 2019-02-19 | | Release date: | 2019-09-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Development of Inhibitors againstMycobacterium abscessustRNA (m1G37) Methyltransferase (TrmD) Using Fragment-Based Approaches.
J.Med.Chem., 62, 2019
|
|
9BG2
 
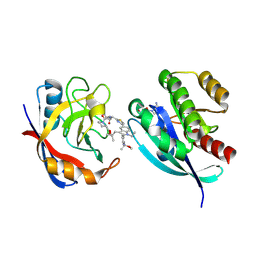 | | Tri-complex of Compound-10, KRAS G12V, and CypA | | Descriptor: | (1S,2R)-N-[(1P,7S,9S,13S,20M)-21-ethyl-20-{2-[(1S)-1-methoxyethyl]pyridin-3-yl}-17,17-dimethyl-8,14-dioxo-15-oxa-4-thia-9,21,27,28-tetraazapentacyclo[17.5.2.1~2,5~.1~9,13~.0~22,26~]octacosa-1(24),2,5(28),19,22,25-hexaen-7-yl]-2-methylcyclopropane-1-carboxamide (non-preferred name), GTPase KRas, MAGNESIUM ION, ... | | Authors: | Tomlinson, A.C.A, Bieder, R, Chen, A, Knox, J.E, Yano, J.K. | | Deposit date: | 2024-04-18 | | Release date: | 2025-03-19 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Discovery of Daraxonrasib (RMC-6236), a Potent and Orally Bioavailable RAS(ON) Multi-selective, Noncovalent Tri-complex Inhibitor for the Treatment of Patients with Multiple RAS-Addicted Cancers.
J.Med.Chem., 68, 2025
|
|
7U8Z
 
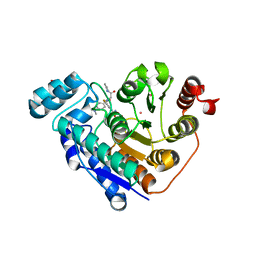 | | Crystal structure of Danio rerio histone deacetylase 6 catalytic domain 2 (CD2) complexed with fluorinated peptoid inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 4-({N-[2-(benzylamino)-2-oxoethyl]-4-(dimethylamino)benzamido}methyl)-3-fluoro-N-hydroxybenzamide, ACETATE ION, ... | | Authors: | Watson, P.R, Cragin, A.D, Christianson, D.W. | | Deposit date: | 2022-03-09 | | Release date: | 2022-11-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Development of Fluorinated Peptoid-Based Histone Deacetylase (HDAC) Inhibitors for Therapy-Resistant Acute Leukemia.
J.Med.Chem., 65, 2022
|
|
5YQO
 
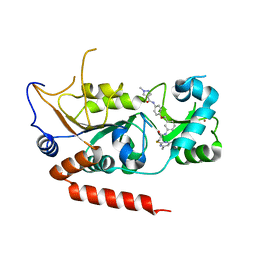 | | Crystal structure of Sirt2 in complex with selective inhibitor L5C | | Descriptor: | N-[4-[[3-[2-(4,6-dimethylpyrimidin-2-yl)sulfanylethanoylamino]phenyl]methoxy]phenyl]-1-methyl-pyrazole-4-carboxamide, NAD-dependent protein deacetylase sirtuin-2, ZINC ION | | Authors: | Wang, H, Yu, Y, Li, G, Chen, Q. | | Deposit date: | 2017-11-07 | | Release date: | 2018-10-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.483 Å) | | Cite: | X-ray crystal structure guided discovery of new selective, substrate-mimicking sirtuin 2 inhibitors that exhibit activities against non-small cell lung cancer cells.
Eur J Med Chem, 155, 2018
|
|
6IBF
 
 | | Crystal structure of human phosphodiesterase 4D2 catalytic domain with inhibitor NPD-417 | | Descriptor: | (4~{a}~{S},8~{a}~{R})-4-[4-methoxy-3-[[2-(trifluoromethyl)phenyl]methoxy]phenyl]-2-(1-thieno[3,2-d]pyrimidin-4-ylpiperidin-4-yl)-4~{a},5,8,8~{a}-tetrahydrophthalazin-1-one, 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Singh, A.K, Brown, D.G. | | Deposit date: | 2018-11-29 | | Release date: | 2019-12-18 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | hPDE4D2 structure with inhibitor NPD-417
To be published
|
|
4CRD
 
 | | Creating novel F1 inhibitors through fragment based lead generation and structure aided drug design | | Descriptor: | COAGULATION FACTOR XI, Methyl N-[4-[5-chloro-2-[[3-[5-chloro-2-(tetrazol-1-yl)phenyl]propanoylamino]methyl]-1H-imidazol-4-yl]phenyl]carbamate, SULFATE ION | | Authors: | Sandmark, J, Oster, L, Fjellstrom, O, Akkaya, S, Beisel, H.G, Eriksson, P.O, Erixon, K, Gustafsson, D, Jurva, U, Kang, D, Karis, D, Knecht, W, Nerme, V, Nilsson, I, Olsson, T, Redzic, A, Roth, R, Tigerstrom, A. | | Deposit date: | 2014-02-26 | | Release date: | 2015-02-11 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Creating Novel Activated Factor Xi Inhibitors Through Fragment Based Lead Generation and Structure Aided Drug Design.
Plos One, 10, 2015
|
|
9BG4
 
 | | Tri-complex of Compound-2, KRAS G12V, and CypA | | Descriptor: | (2R)-N-[(1P,8S,10S,14S,21M)-22-ethyl-21-{2-[(1R)-1-methoxyethyl]pyridin-3-yl}-18,18-dimethyl-9,15-dioxo-16-oxa-10,22,28-triazapentacyclo[18.5.2.1~2,6~.1~10,14~.0~23,27~]nonacosa-1(25),2(29),3,5,20,23,26-heptaen-8-yl]-3-methyl-2-(N-methylacetamido)butanamide (non-preferred name), CHLORIDE ION, GTPase KRas, ... | | Authors: | Tomlinson, A.C.A, Chen, A, Knox, J.E, Yano, J.K. | | Deposit date: | 2024-04-18 | | Release date: | 2025-03-19 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (1.14 Å) | | Cite: | Discovery of Daraxonrasib (RMC-6236), a Potent and Orally Bioavailable RAS(ON) Multi-selective, Noncovalent Tri-complex Inhibitor for the Treatment of Patients with Multiple RAS-Addicted Cancers.
J.Med.Chem., 68, 2025
|
|
6E1Y
 
 | |
6YG2
 
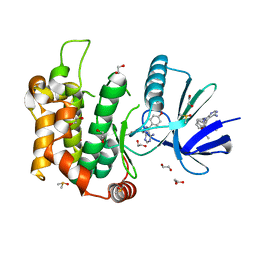 | | Crystal structure of MKK7 (MAP2K7) in complex with ibrutnib, with covalent and allosteric binding modes | | Descriptor: | 1,2-ETHANEDIOL, 1-[(3~{R})-3-[4-azanyl-3-(4-phenoxyphenyl)pyrazolo[3,4-d]pyrimidin-1-yl]piperidin-1-yl]propan-1-one, 1-{(3R)-3-[4-amino-3-(4-phenoxyphenyl)-1H-pyrazolo[3,4-d]pyrimidin-1-yl]piperidin-1-yl}prop-2-en-1-one, ... | | Authors: | Chaikuad, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-03-27 | | Release date: | 2020-08-12 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Catalytic Domain Plasticity of MKK7 Reveals Structural Mechanisms of Allosteric Activation and Diverse Targeting Opportunities.
Cell Chem Biol, 27, 2020
|
|
6E0V
 
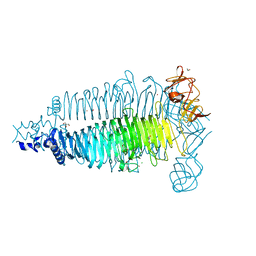 | | Apo crystal structure of the colanidase tailspike protein gp150 of Phage Phi92 | | Descriptor: | 1,2-ETHANEDIOL, Bacteriophage Phi92 gp150, CHLORIDE ION, ... | | Authors: | Leiman, P.G, Browning, C, Gerardy-Schahn, R, Shneider, M.M, Plattner, M, Schwarzer, D. | | Deposit date: | 2018-07-07 | | Release date: | 2019-07-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of the colanidase tailspike protein gp150 of Phage Phi92 complexed with one repeating unit of colanic acid
To Be Published
|
|
7M49
 
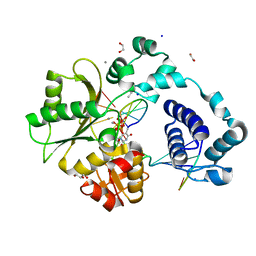 | | DNA Polymerase Lambda, TTP:At Mn2+ Reaction State Ternary Complex, 5 min | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*CP*AP*GP*TP*AP*CP*T)-3'), DNA (5'-D(*CP*GP*GP*CP*AP*GP*TP*AP*CP*TP*G)-3'), ... | | Authors: | Jamsen, J.A, Wilson, S.H. | | Deposit date: | 2021-03-21 | | Release date: | 2022-07-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Watching right and wrong nucleotide insertion captures hidden polymerase fidelity checkpoints.
Nat Commun, 13, 2022
|
|
6DZO
 
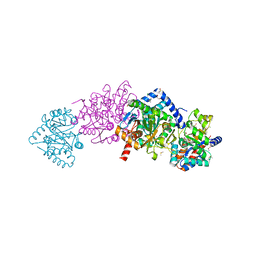 | | Crystal structure of Salmonella typhimurium Tryptophan Synthase mutant beta-Q114A with 2-({[4-(trifluoromethoxy)phenyl]sulfonyl}amino)ethyl dihydrogen phosphate (F9F) at the alpha-site, Cesium ion at the metal coordination site, and (E)-N-({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene)-L-serine at the beta-site | | Descriptor: | (E)-N-({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene)-L-serine, 1,2-ETHANEDIOL, 2-({[4-(TRIFLUOROMETHOXY)PHENYL]SULFONYL}AMINO)ETHYL DIHYDROGEN PHOSPHATE, ... | | Authors: | Hilario, E, Dunn, M.F, Mueller, L.J, Fan, L. | | Deposit date: | 2018-07-05 | | Release date: | 2019-07-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Crystal structure of Salmonella typhimurium Tryptophan Synthase mutant beta-Q114A with 2-({[4-(trifluoromethoxy)phenyl]sulfonyl}amino)ethyl dihydrogen phosphate (F9F) at the alpha-site, Cesium ion at the metal coordination site, and (E)-N-({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene)-L-serine at the beta-site
To be Published
|
|
1L46
 
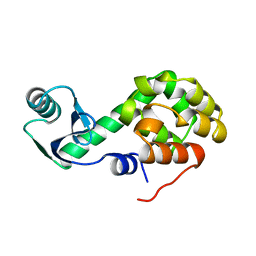 | |
5YPY
 
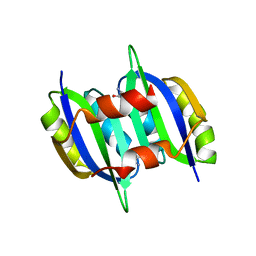 | | Crystal structure of IlvN. Val-1c | | Descriptor: | Acetolactate synthase isozyme 1 small subunit, VALINE | | Authors: | Sarma, S.P, Bansal, A, Schindelin, H, Demeler, B. | | Deposit date: | 2017-11-04 | | Release date: | 2018-09-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.966 Å) | | Cite: | Crystallographic Structures of IlvN·Val/Ile Complexes: Conformational Selectivity for Feedback Inhibition of Aceto Hydroxy Acid Synthases.
Biochemistry, 58, 2019
|
|
1B4N
 
 | | FORMALDEHYDE FERREDOXIN OXIDOREDUCTASE FROM PYROCOCCUS FURIOSUS, COMPLEXED WITH GLUTARATE | | Descriptor: | CALCIUM ION, FORMALDEHYDE FERREDOXIN OXIDOREDUCTASE, GLUTARIC ACID, ... | | Authors: | Hu, Y.L, Faham, S, Roy, R, Adams, M.W.W, Rees, D.C. | | Deposit date: | 1998-12-24 | | Release date: | 1999-05-18 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Formaldehyde ferredoxin oxidoreductase from Pyrococcus furiosus: the 1.85 A resolution crystal structure and its mechanistic implications.
J.Mol.Biol., 286, 1999
|
|
3GOC
 
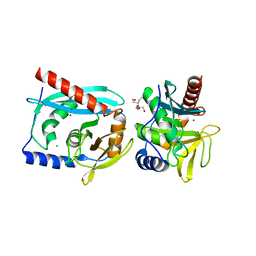 | | Crystal structure of the Endonuclease V (SAV1684) from Streptomyces avermitilis. Northeast Structural Genomics Consortium Target SvR196 | | Descriptor: | 3-(2-hydroxyethyl)-2,2-bis(hydroxymethyl)pentane-1,5-diol, CHLORIDE ION, Endonuclease V, ... | | Authors: | Forouhar, F, Abashidze, M, Hussain, M, Seetharaman, J, Fang, F, Xiao, R, Cunningham, K, Ma, L, Owens, L, Chen, C.X, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-03-18 | | Release date: | 2009-03-31 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of the Endonuclease V (SAV1684) from Streptomyces avermitilis. Northeast Structural Genomics Consortium Target SvR196.
To be Published
|
|
6YPQ
 
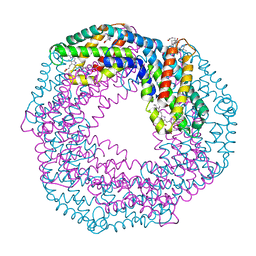 | | Crystal structure of native Phycocyanin from T. elongatus in spacegroup R32 at 1.29 Angstroms | | Descriptor: | C-phycocyanin alpha chain, C-phycocyanin beta chain, GLYCINE, ... | | Authors: | Feiler, C.G, Falke, S, Sarrou, I. | | Deposit date: | 2020-04-16 | | Release date: | 2021-01-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | C-phycocyanin as a highly attractive model system in protein crystallography: unique crystallization properties and packing-diversity screening.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
7M43
 
 | | DNA Polymerase Lambda, TTP:At Ca2+ Ground State Ternary Complex | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, DNA (5'-D(*CP*AP*GP*TP*AP*C)-3'), ... | | Authors: | Jamsen, J.A, Wilson, S.H. | | Deposit date: | 2021-03-21 | | Release date: | 2022-07-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.978 Å) | | Cite: | Watching right and wrong nucleotide insertion captures hidden polymerase fidelity checkpoints.
Nat Commun, 13, 2022
|
|
6QO7
 
 | |
6E1A
 
 | | Menin bound to M-89 | | Descriptor: | (1S,2R)-2-[(4S)-2-methyl-4-{1-[(1-{4-[(pyridin-4-yl)sulfonyl]phenyl}azetidin-3-yl)methyl]piperidin-4-yl}-1,2,3,4-tetrahydroisoquinolin-4-yl]cyclopentyl methylcarbamate, Menin, praseodymium triacetate | | Authors: | Stuckey, J.A. | | Deposit date: | 2018-07-09 | | Release date: | 2019-07-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure-Based Discovery of M-89 as a Highly Potent Inhibitor of the Menin-Mixed Lineage Leukemia (Menin-MLL) Protein-Protein Interaction.
J.Med.Chem., 62, 2019
|
|
5AJY
 
 | | Human PFKFB3 in complex with an indole inhibitor 4 | | Descriptor: | 6-O-phosphono-beta-D-fructofuranose, 6-PHOSPHOFRUCTO-2-KINASE/FRUCTOSE-2,6-BISPHOSPHATASE 3, N-(4-{[3-(1-methyl-1H-pyrazol-4-yl)-1H-indol-5-yl]oxy}phenyl)glycinamide, ... | | Authors: | Boyd, S, Brookfield, J.L, Critchlow, S.E, Cumming, I.A, Curtis, N.J, Debreczeni, J.E, Degorce, S.L, Donald, C, Evans, N.J, Groombridge, S, Hopcroft, P, Jones, N.P, Kettle, J.G, Lamont, S, Lewis, H.J, MacFaull, P, McLoughlin, S.B, Rigoreau, L.J.M, Smith, J.M, St-Gallay, S, Stock, J.K, Wheatley, E.R, Winter, J, Wingfield, J. | | Deposit date: | 2015-02-27 | | Release date: | 2015-04-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Structure-Based Design of Potent and Selective Inhibitors of the Metabolic Kinase Pfkfb3.
J.Med.Chem., 58, 2015
|
|
7M4K
 
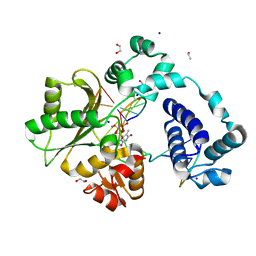 | | DNA Polymerase Lambda, TTPaS:At Ca2+ Ground State Ternary Complex | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, DNA (5'-D(*CP*AP*GP*TP*AP*C)-3'), ... | | Authors: | Jamsen, J.A, Wilson, S.H. | | Deposit date: | 2021-03-21 | | Release date: | 2022-07-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.719 Å) | | Cite: | Watching right and wrong nucleotide insertion captures hidden polymerase fidelity checkpoints.
Nat Commun, 13, 2022
|
|
4GKR
 
 | |
6E1X
 
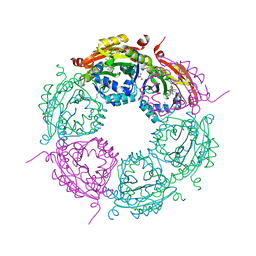 | | Crystal structure of product-bound complex of spermidine/spermine N-acetyltransferase SpeG | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Filippova, E.V, Minasov, G, Kiryukhina, O, Anderson, W.F, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-07-10 | | Release date: | 2019-07-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Crystal structure of product-bound complex of spermidine/spermine N-acetyltransferase SpeG from Vibrio cholerae.
To Be Published
|
|
6QQT
 
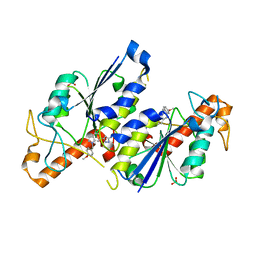 | | Crystal structure of TrmD, a tRNA-(N1G37) methyltransferase, from Mycobacterium abscessus in complex with inhibitor | | Descriptor: | 3-[1-[(4-methoxyphenyl)methyl]indol-6-yl]-1~{H}-pyrazol-5-amine, SULFATE ION, tRNA (guanine-N(1)-)-methyltransferase | | Authors: | Thomas, S.E, Whitehouse, A.J, Coyne, A.G, Abell, C, Mendes, V, Blundell, T.L. | | Deposit date: | 2019-02-19 | | Release date: | 2019-09-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.673 Å) | | Cite: | Development of Inhibitors againstMycobacterium abscessustRNA (m1G37) Methyltransferase (TrmD) Using Fragment-Based Approaches.
J.Med.Chem., 62, 2019
|
|
