7WVZ
 
 | | CalA3_modular PKS_KS-AT-DH-KR | | Descriptor: | Beta-ketoacyl-acyl-carrier-protein synthase I | | Authors: | Wang, J, Wang, Z. | | Deposit date: | 2022-02-12 | | Release date: | 2023-02-22 | | Last modified: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (3.38 Å) | | Cite: | C-N bond formation by a polyketide synthase.
Nat Commun, 14, 2023
|
|
7UOG
 
 | | Isoreticular, interpenetrating co-crystal of Replication Initiator Protein REPE54 and asymmetrical expanded duplex (31mer) containing the cognate REPE54 sequence and an additional G-C rich sequence. | | Descriptor: | DNA (5'-D(AP*CP*CP*TP*GP*TP*GP*AP*CP*AP*AP*AP*TP*TP*GP*CP*CP*CP*TP*CP*AP*GP*AP*CP*CP*CP*GP*CP*CP*TP*T)-3', DNA (5'-D(GP*TP*AP*AP*GP*GP*CP*GP*GP*GP*TP*CP*TP*GP*AP*GP*GP*GP*CP*AP*AP*TP*TP*TP*GP*TP*CP*AP*CP*AP*G)-3', MAGNESIUM ION, ... | | Authors: | Ward, A.R, Snow, C.D. | | Deposit date: | 2022-04-12 | | Release date: | 2023-04-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | Modular Protein-DNA Cocrystals as Precise, Programmable Assembly Scaffolds.
Acs Nano, 17, 2023
|
|
7UV7
 
 | | Isoreticular, interpenetrating co-crystal of Replication Initiator Protein REPE54 and scaffold duplex (21mer) containing the cognate REPE54 sequence and an insert duplex (10mer) with guest TAMRA-labelled thymine and T-A rich sequence. | | Descriptor: | DNA (5'-D(*AP*CP*CP*TP*GP*TP*GP*AP*CP*AP*AP*AP*TP*TP*GP*CP*CP*CP*TP*CP*A)-3'), DNA (5'-D(*GP*AP*CP*GP*GP*TP*AP*AP*TP*T)-3'), DNA (5'-D(*GP*T(TAMRA)P*AP*AP*TP*TP*AP*CP*CP*G)-3'), ... | | Authors: | Orun, A.R, Snow, C.D. | | Deposit date: | 2022-04-29 | | Release date: | 2023-05-17 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3.02 Å) | | Cite: | Modular Protein-DNA Cocrystals as Precise, Programmable Assembly Scaffolds.
Acs Nano, 17, 2023
|
|
7UXY
 
 | | Isoreticular, interpenetrating co-crystal of protein variant Replication Initiator Protein REPE54 (L53G,Q54G,E55G) and symmetrical expanded duplex (31mer) containing the cognate REPE54 sequence and an additional G-C rich sequence. | | Descriptor: | DNA (5'-D(A*CP*CP*CP*GP*GP*AP*CP*CP*TP*GP*TP*P*GP*AP*CP*AP*AP*AP*TP*TP*GP*CP*CP*CP*TP*CP*AP*GP*AP*CP*GP*G)-3'), DNA (5'-D(A*GP*CP*CP*GP*TP*CP*TP*GP*AP*GP*GP*P*GP*CP*AP*AP*TP*TP*TP*GP*TP*CP*AP*CP*AP*GP*GP*TP*CP*CP*GP*G)-3'), MAGNESIUM ION, ... | | Authors: | Orun, A.R, Snow, C.D. | | Deposit date: | 2022-05-06 | | Release date: | 2023-05-17 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Modular Protein-DNA Cocrystals as Precise, Programmable Assembly Scaffolds.
Acs Nano, 17, 2023
|
|
6BA6
 
 | | Solution structure of Rap1b/talin complex | | Descriptor: | Ras-related protein Rap-1b, Talin-1 | | Authors: | Zhu, L, Yang, J, Qin, J. | | Deposit date: | 2017-10-12 | | Release date: | 2017-12-06 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure of Rap1b bound to talin reveals a pathway for triggering integrin activation.
Nat Commun, 8, 2017
|
|
7XI6
 
 | | Crystal structure of C56 from pSSVi | | Descriptor: | C56 | | Authors: | Zhang, Z.F, Ren, Y, Chen, Y.Y, Zhang, X.W, Dong, Y.H, Gong, Y, Cao, P, Huang, L. | | Deposit date: | 2022-04-12 | | Release date: | 2023-04-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Characterization of the AbrB-like protein C56 conserved in a novel family of integrated genetic elements in Sulfolobales
To Be Published
|
|
4YDH
 
 | | The structure of human FMNL1 N-terminal domains bound to Cdc42 | | Descriptor: | Cell division control protein 42 homolog, Formin-like protein 1, MAGNESIUM ION, ... | | Authors: | Kuhn, S, Anand, K, Geyer, M. | | Deposit date: | 2015-02-22 | | Release date: | 2015-05-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | The structure of FMNL2-Cdc42 yields insights into the mechanism of lamellipodia and filopodia formation.
Nat Commun, 6, 2015
|
|
7XL6
 
 | |
4YC7
 
 | |
7S0Y
 
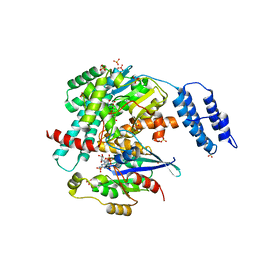 | | Structures of TcdB in complex with Cdc42 | | Descriptor: | Cell division control protein 42 homolog, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Zheng, L, Rongsheng, J, Peng, C. | | Deposit date: | 2021-08-31 | | Release date: | 2021-09-08 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Structural basis for selective modification of Rho and Ras GTPases by Clostridioides difficile toxin B.
Sci Adv, 7, 2021
|
|
4ZDG
 
 | |
7Y0V
 
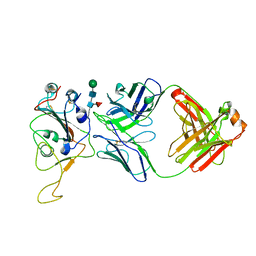 | | The co-crystal structure of BA.1-RBD with Fab-5549 | | Descriptor: | 5549-Fab, Spike protein S1, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Xiao, J.Y, Zhang, Y. | | Deposit date: | 2022-06-06 | | Release date: | 2022-09-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Rational identification of potent and broad sarbecovirus-neutralizing antibody cocktails from SARS convalescents.
Cell Rep, 41, 2022
|
|
7Y1G
 
 | | Crystal structure of human PRKACA complexed with DS01080522 | | Descriptor: | 1-chloranyl-~{N}-[(~{S})-(3-chloranyl-4-cyano-phenyl)-[(2~{R},4~{S})-4-oxidanylpyrrolidin-2-yl]methyl]-7-methoxy-isoquinoline-6-carboxamide, ZINC ION, cAMP-dependent protein kinase catalytic subunit alpha | | Authors: | Suzuki, M, Ubukata, O, Toyoda, A. | | Deposit date: | 2022-06-08 | | Release date: | 2022-09-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Novel protein kinase cAMP-Activated Catalytic Subunit Alpha (PRKACA) inhibitor shows anti-tumor activity in a fibrolamellar hepatocellular carcinoma model.
Biochem.Biophys.Res.Commun., 621, 2022
|
|
7Y5T
 
 | | CryoEM structure of PS1-containing gamma-secretase in complex with MRK-560 | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Guo, X, Wang, Y, Zhou, J, Jin, C, Wang, J, Jia, B, Jing, D, Yan, C, Lei, J, Zhou, R, Shi, Y. | | Deposit date: | 2022-06-17 | | Release date: | 2022-11-02 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Molecular basis for isoform-selective inhibition of presenilin-1 by MRK-560.
Nat Commun, 13, 2022
|
|
7SH2
 
 | | Structure of the yeast Rad24-RFC loader bound to DNA and the open 9-1-1 clamp | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Checkpoint protein RAD24, Crick strand, ... | | Authors: | Zheng, F, Georgescu, R, Yao, Y.N, O'Donnell, M.E, Li, H. | | Deposit date: | 2021-10-07 | | Release date: | 2022-03-23 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.23 Å) | | Cite: | DNA is loaded through the 9-1-1 DNA checkpoint clamp in the opposite direction of the PCNA clamp.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7SGZ
 
 | | Structure of the yeast Rad24-RFC loader bound to DNA and the closed 9-1-1 clamp | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Checkpoint protein RAD24, Crick strand, ... | | Authors: | Zheng, F, Georgescu, R, Yao, Y.N, O'Donnell, M.E, Li, H. | | Deposit date: | 2021-10-07 | | Release date: | 2022-03-23 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.17 Å) | | Cite: | DNA is loaded through the 9-1-1 DNA checkpoint clamp in the opposite direction of the PCNA clamp.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7RY0
 
 | | human Hsp90_MC domain structure | | Descriptor: | DIMETHYL SULFOXIDE, GLYCEROL, Heat shock protein HSP 90-alpha | | Authors: | Peng, S, Deng, J, Matts, R. | | Deposit date: | 2021-08-24 | | Release date: | 2022-12-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis of the key residue W320 responsible for Hsp90 conformational change.
J.Biomol.Struct.Dyn., 2022
|
|
7UR0
 
 | | Isoreticular, interpenetrating co-crystal of Replication Initiator Protein REPE54 and scaffold-insert duplexes (21mer and 10mer) containing the cognate REPE54 sequence and an additional T-A rich sequence, respectively. | | Descriptor: | DNA (5'-D(*AP*AP*CP*CP*TP*GP*TP*GP*AP*CP*AP*AP*AP*TP*TP*GP*CP*CP*CP*TP*CP*A)-3'), DNA (5'-D(A*GP*AP*CP*GP*GP*TP*AP*AP*TP*T)-3'), DNA (5'-D(A*GP*TP*AP*AP*TP*TP*AP*CP*CP*G)-3'), ... | | Authors: | Orun, A.R, Snow, C.D. | | Deposit date: | 2022-04-20 | | Release date: | 2023-04-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Modular Protein-DNA Cocrystals as Precise, Programmable Assembly Scaffolds.
Acs Nano, 17, 2023
|
|
8IFC
 
 | | Arbekacin-bound E.coli 70S ribosome in the PURE system | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Tomono, J, Asano, K, Chiashi, T, Tanaka, Y, Yokoyama, T. | | Deposit date: | 2023-02-17 | | Release date: | 2024-02-14 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Direct visualization of ribosomes in the cell-free system revealed the functional evolution of aminoglycoside.
J.Biochem., 175, 2024
|
|
8IFB
 
 | | Dibekacin-bound E.coli 70S ribosome in the PURE system | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Tomono, J, Asano, K, Chiashi, T, Tanaka, Y, Yokoyama, T. | | Deposit date: | 2023-02-17 | | Release date: | 2024-02-14 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.43 Å) | | Cite: | Direct visualization of ribosomes in the cell-free system revealed the functional evolution of aminoglycoside.
J.Biochem., 175, 2024
|
|
8GI6
 
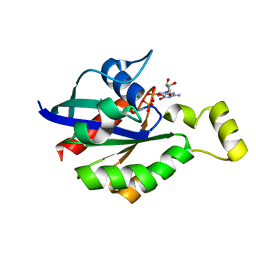 | | Crystal structure of RhoA mutant L69R complexed with GDP | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Transforming protein RhoA | | Authors: | Chen, X, Qian, X, Chandravanshi, M, Lowy, D.R, Walters, K.J. | | Deposit date: | 2023-03-13 | | Release date: | 2024-03-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Ras-like GTPases mutant structures
To be published
|
|
8GI3
 
 | | Crystal structure of RhoA mutant L69P complexed with GDP | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Transforming protein RhoA | | Authors: | Chen, X, Qian, X, Chandravanshi, M, Lowy, D.R, Walters, K.J. | | Deposit date: | 2023-03-13 | | Release date: | 2024-03-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Ras-like GTPases mutants structure
To be published
|
|
5AY6
 
 | |
4R89
 
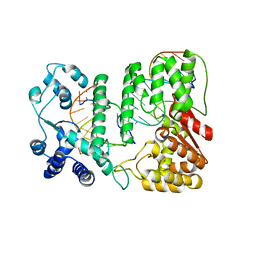 | | Crystal structure of paFAN1 - 5' flap DNA complex with Manganase | | Descriptor: | DNA (5'-D(P*AP*CP*CP*AP*GP*AP*CP*AP*CP*AP*CP*AP*TP*TP*C)-3'), DNA (5'-D(P*GP*AP*AP*TP*GP*TP*GP*TP*GP*TP*CP*TP*CP*AP*AP*TP*CP*CP*CP*AP*AP*C)-3'), DNA (5'-D(P*GP*TP*TP*GP*GP*GP*AP*TP*TP*G)-3'), ... | | Authors: | Cho, Y, Gwon, G.H, Kim, Y.R. | | Deposit date: | 2014-08-30 | | Release date: | 2014-10-29 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (4.002 Å) | | Cite: | Crystal structure of a Fanconi anemia-associated nuclease homolog bound to 5' flap DNA: basis of interstrand cross-link repair by FAN1
Genes Dev., 28, 2014
|
|
4R8A
 
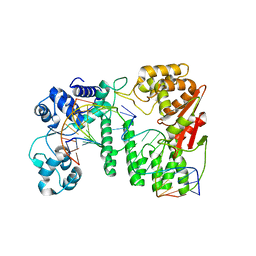 | | Crystal structure of paFAN1 - 5' flap DNA complex | | Descriptor: | DNA (5'-D(P*AP*CP*CP*AP*GP*AP*CP*AP*CP*AP*CP*AP*TP*TP*C)-3'), DNA (5'-D(P*GP*AP*AP*TP*GP*TP*GP*TP*GP*TP*CP*TP*CP*AP*AP*TP*CP*CP*CP*AP*A)-3'), DNA (5'-D(P*GP*TP*TP*GP*GP*GP*AP*TP*TP*G)-3'), ... | | Authors: | Cho, Y, Gwon, G.H, Kim, Y.R. | | Deposit date: | 2014-08-30 | | Release date: | 2014-10-29 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of a Fanconi anemia-associated nuclease homolog bound to 5' flap DNA: basis of interstrand cross-link repair by FAN1
Genes Dev., 28, 2014
|
|
