1KK3
 
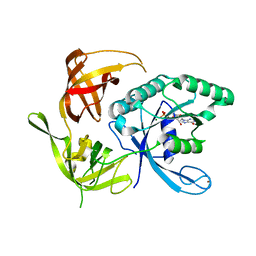 | | Structure of the wild-type large gamma subunit of initiation factor eIF2 from Pyrococcus abyssi complexed with GDP-Mg2+ | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ZINC ION, ... | | Authors: | Schmitt, E, Blanquet, S, Mechulam, Y. | | Deposit date: | 2001-12-06 | | Release date: | 2002-04-10 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The large subunit of initiation factor aIF2 is a close structural homologue of elongation factors.
EMBO J., 21, 2002
|
|
1KKF
 
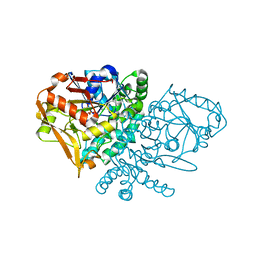 | | Complex of E. coli Adenylosuccinate Synthetase with IMP, Hadacidin, Pyrophosphate, and Mg | | Descriptor: | Adenylosuccinate Synthetase, DIPHOSPHATE, HADACIDIN, ... | | Authors: | Hou, Z, Wang, W, Fromm, H.J, Honzatko, R.B. | | Deposit date: | 2001-12-07 | | Release date: | 2002-03-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | IMP Alone Organizes the Active Site of Adenylosuccinate Synthetase from Escherichia coli.
J.Biol.Chem., 277, 2002
|
|
7A3U
 
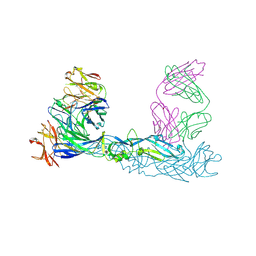 | | Crystal structure of Zika virus envelope glycoprotein in complex with the divalent F(ab')2 fragment of the broadly neutralizing human antibody EDE1 C10 | | Descriptor: | EDE1 C10 antibody divalent F(ab')2 fragment, EDE1 C10 divalent F(ab')2 fragment, Envelope protein | | Authors: | Sharma, A, Vaney, M.C, Guardado-Calvo, P, Duquerroy, S, Rouvinski, A, Rey, F.A. | | Deposit date: | 2020-08-18 | | Release date: | 2021-12-08 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The epitope arrangement on flavivirus particles contributes to Mab C10's extraordinary neutralization breadth across Zika and dengue viruses.
Cell, 184, 2021
|
|
1KKQ
 
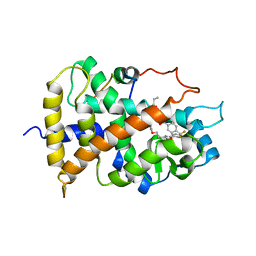 | | Crystal structure of the human PPAR-alpha ligand-binding domain in complex with an antagonist GW6471 and a SMRT corepressor motif | | Descriptor: | N-((2S)-2-({(1Z)-1-METHYL-3-OXO-3-[4-(TRIFLUOROMETHYL) PHENYL]PROP-1-ENYL}AMINO)-3-{4-[2-(5-METHYL-2-PHENYL-1,3-OXAZOL-4-YL)ETHOXY]PHENYL}PROPYL)PROPANAMIDE, NUCLEAR RECEPTOR CO-REPRESSOR 2, PEROXISOME PROLIFERATOR ACTIVATED RECEPTOR | | Authors: | Xu, H.E, Stanley, T.B, Montana, V.G, Lambert, M.H, Shearer, B.G, Cobb, J.E, McKee, D.D, Galardi, C.M, Nolte, R.T, Parks, D.J. | | Deposit date: | 2001-12-10 | | Release date: | 2002-02-20 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis for antagonist-mediated recruitment of nuclear co-repressors by PPARalpha.
Nature, 415, 2002
|
|
1KL9
 
 | |
1KN7
 
 | | Solution structure of the tandem inactivation domain (residues 1-75) of potassium channel RCK4 (Kv1.4) | | Descriptor: | VOLTAGE-GATED POTASSIUM CHANNEL PROTEIN KV1.4 | | Authors: | Wissmann, R, Bildl, W, Oliver, D, Beyermann, M, Kalbitzer, H.R, Bentrop, D, Fakler, B. | | Deposit date: | 2001-12-18 | | Release date: | 2003-05-06 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution Structure and Function of the "Tandem Inactivation Domain" of the Neuronal A-type
Potassium Channel Kv1.4
J.Biol.Chem., 278, 2003
|
|
1KOE
 
 | | ENDOSTATIN | | Descriptor: | ENDOSTATIN | | Authors: | Hohenester, E, Sasaki, T, Olsen, B.R, Timpl, R. | | Deposit date: | 1998-01-22 | | Release date: | 1999-02-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of the angiogenesis inhibitor endostatin at 1.5 A resolution.
EMBO J., 17, 1998
|
|
1KOP
 
 | | NEISSERIA GONORRHOEAE CARBONIC ANHYDRASE | | Descriptor: | AZIDE ION, BETA-MERCAPTOETHANOL, CARBONIC ANHYDRASE, ... | | Authors: | Huang, S, Xue, Y, Chirica, L, Lindskog, S, Jonsson, B.-H. | | Deposit date: | 1998-03-22 | | Release date: | 1998-12-09 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of carbonic anhydrase from Neisseria gonorrhoeae and its complex with the inhibitor acetazolamide.
J.Mol.Biol., 283, 1998
|
|
6VT3
 
 | | Naegleria gruberi RNA ligase K326A mutant apo | | Descriptor: | RNA Ligase | | Authors: | Unciuleac, M.C, Goldgur, Y, Shuman, S. | | Deposit date: | 2020-02-12 | | Release date: | 2020-04-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.844 Å) | | Cite: | Caveat mutator: alanine substitutions for conserved amino acids in RNA ligase elicit unexpected rearrangements of the active site for lysine adenylylation.
Nucleic Acids Res., 48, 2020
|
|
1KP2
 
 | |
6VTD
 
 | | Naegleria gruberi RNA ligase R149A mutant with ATP and Mn | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MANGANESE (II) ION, RNA Ligase | | Authors: | Unciuleac, M.C, Goldgur, Y, Shuman, S. | | Deposit date: | 2020-02-12 | | Release date: | 2020-04-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Caveat mutator: alanine substitutions for conserved amino acids in RNA ligase elicit unexpected rearrangements of the active site for lysine adenylylation.
Nucleic Acids Res., 48, 2020
|
|
1KLX
 
 | |
1KM0
 
 | |
1KMI
 
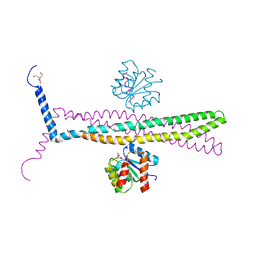 | | CRYSTAL STRUCTURE OF AN E.COLI CHEMOTAXIS PROTEIN, CHEZ | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, BICINE, Chemotaxis protein cheY, ... | | Authors: | Zhao, R, Collins, E.J, Bourret, R.B, Silversmith, R.E. | | Deposit date: | 2001-12-16 | | Release date: | 2002-07-24 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure and catalytic mechanism of the E. coli chemotaxis phosphatase CheZ.
Nat.Struct.Biol., 9, 2002
|
|
1KMO
 
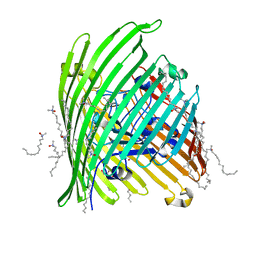 | | Crystal structure of the Outer Membrane Transporter FecA | | Descriptor: | HEPTANE-1,2,3-TRIOL, Iron(III) dicitrate transport protein fecA, LAURYL DIMETHYLAMINE-N-OXIDE | | Authors: | Ferguson, A.D, Chakraborty, R, Smith, B.S, Esser, L, van der Helm, D, Deisenhofer, J. | | Deposit date: | 2001-12-17 | | Release date: | 2002-03-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis of gating by the outer membrane transporter FecA.
Science, 295, 2002
|
|
1KN5
 
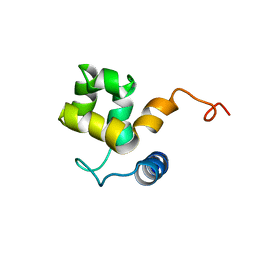 | | SOLUTION STRUCTURE OF ARID DOMAIN OF ADR6 FROM SACCHAROMYCES CEREVISIAE | | Descriptor: | Transcription regulatory protein ADR6 | | Authors: | Tu, X, Wu, J, Xu, Y, Shi, Y. | | Deposit date: | 2001-12-18 | | Release date: | 2002-07-17 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | 1H, 13C and 15N resonance assignments and secondary structure of ADR6 DNA-binding domain.
J.Biomol.Nmr, 21, 2001
|
|
1KND
 
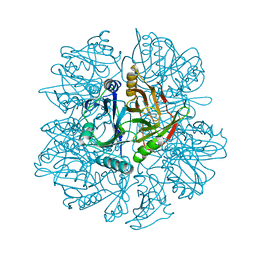 | | Crystal Structure of 2,3-dihydroxybiphenyl 1,2-dioxygenase Complexed with Catechol under Anaerobic Condition | | Descriptor: | 2,3-DIHYDROXYBIPHENYL 1,2-DIOXYGENASE, CATECHOL, FE (II) ION, ... | | Authors: | Han, S, Bolin, J.T. | | Deposit date: | 2001-12-18 | | Release date: | 2002-03-27 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Molecular basis for the stabilization and inhibition of 2, 3-dihydroxybiphenyl 1,2-dioxygenase by t-butanol.
J.Biol.Chem., 273, 1998
|
|
1KNP
 
 | |
1KPG
 
 | | Crystal Structure of mycolic acid cyclopropane synthase CmaA1 complexed with SAH and CTAB | | Descriptor: | CARBONATE ION, CETYL-TRIMETHYL-AMMONIUM, CYCLOPROPANE-FATTY-ACYL-PHOSPHOLIPID SYNTHASE 1, ... | | Authors: | Huang, C.-C, Smith, C.V, Jacobs Jr, W.R, Glickman, M.S, Sacchettini, J.C, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2001-12-30 | | Release date: | 2002-01-11 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of mycolic acid cyclopropane synthases from Mycobacterium tuberculosis
J.Biol.Chem., 277, 2002
|
|
1KQ7
 
 | | E315Q Mutant Form of Fumarase C from E.coli | | Descriptor: | CITRIC ACID, D-MALATE, FUMARATE HYDRATASE CLASS II | | Authors: | Weaver, T.M, Estevez, M, Skarda, J, Spencer, J. | | Deposit date: | 2002-01-04 | | Release date: | 2002-08-23 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | X-ray crystallographic and kinetic correlation of a clinically observed human fumarase mutation.
Protein Sci., 11, 2002
|
|
6VT5
 
 | |
1KO3
 
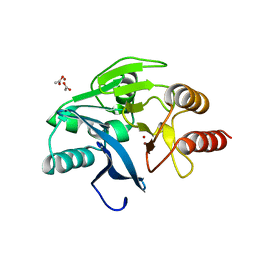 | | VIM-2, a Zn-beta-lactamase from Pseudomonas aeruginosa with Cys221 reduced | | Descriptor: | ACETATE ION, CHLORIDE ION, HYDROXIDE ION, ... | | Authors: | Garcia-Saez, I, Docquier, J.-D, Rossolini, G.M, Dideberg, O. | | Deposit date: | 2001-12-20 | | Release date: | 2003-09-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | The three-dimensional structure of VIM-2, a Zn-beta-lactamase from Pseudomonas aeruginosa in its reduced and oxidised form
J.Mol.Biol., 375, 2008
|
|
6VTF
 
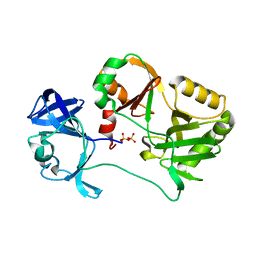 | | Naegleria gruberi RNA ligase with PPi | | Descriptor: | PYROPHOSPHATE 2-, RNA ligase | | Authors: | Unciuleac, M.C, Goldgur, Y, Shuman, S. | | Deposit date: | 2020-02-12 | | Release date: | 2020-04-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Caveat mutator: alanine substitutions for conserved amino acids in RNA ligase elicit unexpected rearrangements of the active site for lysine adenylylation.
Nucleic Acids Res., 48, 2020
|
|
1KOF
 
 | | Crystal structure of gluconate kinase | | Descriptor: | Gluconate kinase, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER | | Authors: | Kraft, L, Sprenger, G.A, Lindqvist, Y. | | Deposit date: | 2001-12-20 | | Release date: | 2002-05-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Conformational changes during the catalytic cycle of gluconate kinase as revealed by X-ray crystallography.
J.Mol.Biol., 318, 2002
|
|
1KOS
 
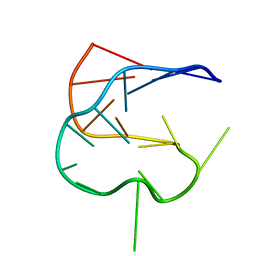 | | SOLUTION NMR STRUCTURE OF AN ANALOG OF THE YEAST TRNA PHE T STEM LOOP CONTAINING RIBOTHYMIDINE AT ITS NATURALLY OCCURRING POSITION | | Descriptor: | 5'-R(*CP*UP*GP*UP*GP*(5MU)P*UP*CP*GP*AP*UP*(CH)P*CP*AP*CP*AP*G)- 3' | | Authors: | Koshlap, K.M, Guenther, R, Sochacka, E, Malkiewicz, A, Agris, P.F. | | Deposit date: | 1999-05-03 | | Release date: | 1999-10-22 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | A distinctive RNA fold: the solution structure of an analogue of the yeast tRNAPhe T Psi C domain.
Biochemistry, 38, 1999
|
|
