4R7H
 
 | | Crystal structure of FMS KINASE domain with a small molecular inhibitor, PLX3397 | | Descriptor: | 5-[(5-chloro-1H-pyrrolo[2,3-b]pyridin-3-yl)methyl]-N-{[6-(trifluoromethyl)pyridin-3-yl]methyl}pyridin-2-amine, Macrophage colony-stimulating factor 1 receptor | | Authors: | Zhang, Y, Zhang, K, Zhang, C. | | Deposit date: | 2014-08-27 | | Release date: | 2015-08-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.8001 Å) | | Cite: | Structure-Guided Blockade of CSF1R Kinase in Tenosynovial Giant-Cell Tumor.
N Engl J Med, 373, 2015
|
|
8ROY
 
 | | Structure of the human DDB1-DDA1-DCAF15 E3 ubiquitin ligase bound to compound furan 24 | | Descriptor: | 1-[5-[[3,4-bis(chloranyl)-1~{H}-indol-7-yl]sulfamoyl]-3-methyl-furan-2-yl]carbonyl-~{N}-methyl-piperidine-4-carboxamide, DDB1- and CUL4-associated factor 15, DET1- and DDB1-associated protein 1, ... | | Authors: | Shilliday, F, Lucas, S.C.C, Richter, M, Michaelides, I.N, Fusani, L. | | Deposit date: | 2024-01-12 | | Release date: | 2024-04-03 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Optimization of Potent Ligands for the E3 Ligase DCAF15 and Evaluation of Their Use in Heterobifunctional Degraders.
J.Med.Chem., 67, 2024
|
|
9GHN
 
 | | Structure of SARS-CoV-2 Main Protease (Mpro) with mutation of Q256A | | Descriptor: | 1,2-ETHANEDIOL, 3C-like proteinase nsp5, CHLORIDE ION, ... | | Authors: | Creon, A, Scheer, T.E.S, Lane, T.J, Rahmani Mashhour, A, Guenther, S, Reinke, P.Y.A, Meents, A, Chapman, H.N. | | Deposit date: | 2024-08-15 | | Release date: | 2025-09-03 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure of SARS-CoV-2 Main Protease (Mpro) with mutation of Q256A
To Be Published
|
|
6TLO
 
 | | HUMAN CK2 KINASE ALPHA SUBUNIT IN COMPLEX WITH THE ATP-COMPETITIVE INHIBITOR 4,5,6-TRIBROMOBENZOTRIAZOLE | | Descriptor: | 5,6,7-tris(bromanyl)-1~{H}-benzotriazole, CHLORIDE ION, Casein kinase II subunit alpha, ... | | Authors: | Czapinska, H, Piasecka, A, Winiewska-Szajewska, M, Bochtler, M, Poznanski, J. | | Deposit date: | 2019-12-03 | | Release date: | 2020-12-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Halogen Atoms in the Protein-Ligand System. Structural and Thermodynamic Studies of the Binding of Bromobenzotriazoles by the Catalytic Subunit of Human Protein Kinase CK2.
J.Phys.Chem.B, 125, 2021
|
|
6YQN
 
 | | Crystal structure of the first bromodomain of human BRD4 in complex with the dual inhibitor TW9 | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, ~{N}-(2-aminophenyl)-4-[2-[(9~{S})-7-(4-chlorophenyl)-4,5,13-trimethyl-3-thia-1,8,11,12-tetrazatricyclo[8.3.0.0^{2,6}]t rideca-2(6),4,7,10,12-pentaen-9-yl]ethanoylamino]benzamide | | Authors: | Joerger, A.C, Balourdas, D.I, Weiser, T, Chatterjee, D, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-04-17 | | Release date: | 2020-05-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Characterization of a dual BET/HDAC inhibitor for treatment of pancreatic ductal adenocarcinoma.
Int.J.Cancer, 147, 2020
|
|
8VXZ
 
 | | Mango II bound to 365A-084 | | Descriptor: | 3-[2-(benzylamino)-2-oxoethyl]-2-[(E)-(1-methylquinolin-4(1H)-ylidene)methyl]-1,3-benzothiazol-3-ium, Chains: A,B,C, POTASSIUM ION | | Authors: | Passalacqua, L.F.M, Ferre-D'Amare, A.R, Schneekloth, J.S. | | Deposit date: | 2024-02-06 | | Release date: | 2025-02-19 | | Last modified: | 2025-10-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure-informed design of an ultrabright RNA-activated fluorophore.
Nat.Chem., 17, 2025
|
|
8VY1
 
 | | Mango II bound to 365A-088 | | Descriptor: | 3-(2-{[(2-fluorophenyl)methyl]amino}-2-oxoethyl)-2-[(E)-(1-methylquinolin-4(1H)-ylidene)methyl]-1,3-benzothiazol-3-ium, Chains: A,B,C, POTASSIUM ION | | Authors: | Passalacqua, L.F.M, Ferre-D'Amare, A.R, Schneekloth, J.S. | | Deposit date: | 2024-02-06 | | Release date: | 2025-02-19 | | Last modified: | 2025-10-08 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure-informed design of an ultrabright RNA-activated fluorophore.
Nat.Chem., 17, 2025
|
|
7Y0L
 
 | | SulE-S209A | | Descriptor: | Alpha/beta fold hydrolase, GLYCEROL, METHYL 2-[({[(4-METHOXY-6-METHYL-1,3,5-TRIAZIN-2-YL)AMINO]CARBONYL}AMINO)SULFONYL]BENZOATE | | Authors: | Liu, B, Ran, T, He, J, Wang, W. | | Deposit date: | 2022-06-05 | | Release date: | 2023-08-02 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | Crystal structures of herbicide-detoxifying esterase reveal a lid loop affecting substrate binding and activity.
Nat Commun, 14, 2023
|
|
5DD4
 
 | | Apo structure of transcriptional factor AraR from Bacteroides thetaiotaomicron VPI | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, TRANSCRIPTIONAL REGULATOR AraR | | Authors: | Chang, C, Tesar, C, Rodionov, D, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2015-08-24 | | Release date: | 2015-09-09 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | A novel transcriptional regulator of L-arabinose utilization in human gut bacteria.
Nucleic Acids Res., 43, 2015
|
|
8GC1
 
 | |
8VXX
 
 | | Mango II bound to 365A-061 | | Descriptor: | 3-[2-({[(6P)-2-fluoro-6-(1H-pyrazol-1-yl)phenyl]methyl}amino)-2-oxoethyl]-2-[(E)-(1-methylquinolin-4(1H)-ylidene)methyl]-1,3-benzothiazol-3-ium, Chains: A,B,C, POTASSIUM ION | | Authors: | Passalacqua, L.F.M, Ferre-D'Amare, A.R, Schneekloth, J.S. | | Deposit date: | 2024-02-06 | | Release date: | 2025-02-19 | | Last modified: | 2025-10-08 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure-informed design of an ultrabright RNA-activated fluorophore.
Nat.Chem., 17, 2025
|
|
6PXX
 
 | | Class D beta-lactamase in complex with beta-lactam antibiotic | | Descriptor: | (2~{S},3~{R})-3-methyl-2-[(2~{S},3~{R})-3-oxidanyl-1-oxidanylidene-butan-2-yl]-4-[(3~{S},5~{S})-5-[(sulfamoylamino)meth yl]pyrrolidin-3-yl]sulfanyl-3,4-dihydro-2~{H}-pyrrole-5-carboxylic acid, 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | van den Akker, F, Kumar, V. | | Deposit date: | 2019-07-28 | | Release date: | 2019-10-09 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural Analysis of The OXA-48 Carbapenemase Bound to A "Poor" Carbapenem Substrate, Doripenem.
Antibiotics, 8, 2019
|
|
6YBF
 
 | |
7Y94
 
 | |
6TLV
 
 | | HUMAN CK2 KINASE ALPHA SUBUNIT IN COMPLEX WITH THE ATP-COMPETITIVE INHIBITOR 5-BROMOBENZOTRIAZOLE | | Descriptor: | 6-bromanyl-1~{H}-benzotriazole, CHLORIDE ION, Casein kinase II subunit alpha, ... | | Authors: | Czapinska, H, Piasecka, A, Winiewska-Szajewska, M, Bochtler, M, Poznanski, J. | | Deposit date: | 2019-12-03 | | Release date: | 2020-12-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Halogen Atoms in the Protein-Ligand System. Structural and Thermodynamic Studies of the Binding of Bromobenzotriazoles by the Catalytic Subunit of Human Protein Kinase CK2.
J.Phys.Chem.B, 125, 2021
|
|
4R3J
 
 | | Structure of a putative peptidoglycan glycosyltransferase from Atopobium parvulum in complex with cefapirin | | Descriptor: | (2R)-2-[(1R)-1-(acetylamino)-2-oxoethyl]-5-methyl-3,6-dihydro-2H-1,3-thiazine-4-carboxylic acid, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Filippova, E.V, Minasov, G, Kiryukhina, O, Clancy, S, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-08-15 | | Release date: | 2014-09-17 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Structure of a putative peptidoglycan glycosyltransferase from Atopobium parvulum in complex with cefapirin
To be Published
|
|
5GU0
 
 | | Crystal structure of Au.CL-apo-E45C/R52C-rHLFr | | Descriptor: | 1,2-ETHANEDIOL, CADMIUM ION, Ferritin light chain, ... | | Authors: | Maity, B, Abe, S, Ueno, T. | | Deposit date: | 2016-08-24 | | Release date: | 2017-03-29 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Observation of gold sub-nanocluster nucleation within a crystalline protein cage
Nat Commun, 8, 2017
|
|
5GMM
 
 | | Crystal structure of human Carbonic anhydrase I in complex with polmacoxib | | Descriptor: | 4-[3-(3-fluorophenyl)-5,5-dimethyl-4-oxidanylidene-furan-2-yl]benzenesulfonamide, Carbonic anhydrase 1, ZINC ION | | Authors: | Kim, H.T, Hwang, K.Y. | | Deposit date: | 2016-07-14 | | Release date: | 2017-05-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.003 Å) | | Cite: | Structural insight into the inhibition of carbonic anhydrase by the COX-2-selective inhibitor polmacoxib (CG100649).
Biochem. Biophys. Res. Commun., 478, 2016
|
|
9GUD
 
 | | SARS-CoV-2 methyltransferase nsp10-16 in complex with SAM and theophylline derivative LAS 54570922 | | Descriptor: | (3~{S})-3-azanyl-4-[(3~{R},4~{R},6~{S})-3-[1,3-dimethyl-2,6-bis(oxidanylidene)purin-7-yl]-4-methyl-4,6-bis(oxidanyl)azepan-1-yl]-4-oxidanylidene-butanoic acid, 1,2-ETHANEDIOL, 2'-O-methyltransferase nsp16, ... | | Authors: | Kremling, V, Sprenger, J, Oberthuer, D, Kiene, A. | | Deposit date: | 2024-09-19 | | Release date: | 2024-12-04 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structures of SARS-CoV-2 methyltransferase nsp10-16 with Cap0-site binders
To Be Published
|
|
7LP4
 
 | | Structure of Nedd4L WW3 domain | | Descriptor: | E3 ubiquitin-protein ligase NEDD4-like | | Authors: | Alam, S.L, Alian, A, Thompson, T, Rheinemann, L, Volkman, B.F, Peterson, F.C, Sundquist, W.I. | | Deposit date: | 2021-02-11 | | Release date: | 2021-07-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Interactions between AMOT PPxY motifs and NEDD4L WW domains function in HIV-1 release.
J.Biol.Chem., 297, 2021
|
|
9GS4
 
 | | SARS-CoV-2 methyltransferase nsp10-16 in complex with SAM and theophylline derivative LAS 54571130 | | Descriptor: | 1,2-ETHANEDIOL, 2'-O-methyltransferase nsp16, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ... | | Authors: | Kremling, V, Sprenger, J, Oberthuer, D, Kiene, A. | | Deposit date: | 2024-09-13 | | Release date: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of SARS-CoV-2 methyltransferase nsp10-16 with Cap0-site binders
To Be Published
|
|
9QNZ
 
 | | Streptavidin 112Y-121W-124F with a thiophenol cofactor as artificial hydrogen atom transferase | | Descriptor: | 1,2-ETHANEDIOL, 5-[(3~{a}~{S},4~{S},6~{a}~{R})-2-oxidanylidene-1,3,3~{a},4,6,6~{a}-hexahydrothieno[3,4-d]imidazol-4-yl]-~{N}-(3-sulfanylphenyl)pentanamide, ACETATE ION, ... | | Authors: | Zhang, K, Ward, T.R. | | Deposit date: | 2025-03-25 | | Release date: | 2025-11-12 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | An Asymmetric Hydrogen Atom Transferase with an Abiological Thiophenol Cofactor.
J.Am.Chem.Soc., 2025
|
|
6QAG
 
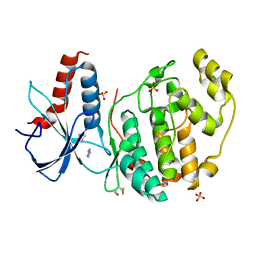 | | ERK2 mini-fragment binding | | Descriptor: | 1~{H}-1,2,3-triazole, Mitogen-activated protein kinase 1, SULFATE ION | | Authors: | O'Reilly, M, Cleasby, A, Davies, T.G, Hall, R, Ludlow, F, Murray, C.W, Tisi, D, Jhoti, H. | | Deposit date: | 2018-12-19 | | Release date: | 2019-03-27 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Crystallographic screening using ultra-low-molecular-weight ligands to guide drug design.
Drug Discov Today, 24, 2019
|
|
4R92
 
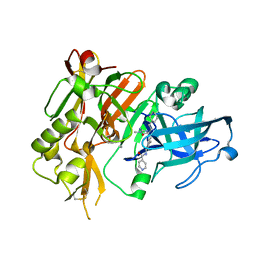 | | BACE-1 in complex with (R)-4-(2-cyclohexylethyl)-4-(((1S,3R)-3-(isonicotinamido)cyclohexyl)methyl)-1-methyl-5-oxoimidazolidin-2-iminium | | Descriptor: | Beta-secretase 1, L(+)-TARTARIC ACID, N-[(1R,3S)-3-{[(2E,4R)-4-(2-cyclohexylethyl)-2-imino-1-methyl-5-oxoimidazolidin-4-yl]methyl}cyclohexyl]pyridine-4-carboxamide | | Authors: | Orth, P, Strickland, C, Caldwell, J.P. | | Deposit date: | 2014-09-03 | | Release date: | 2014-11-05 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Discovery of potent iminoheterocycle BACE1 inhibitors.
Bioorg.Med.Chem.Lett., 24, 2014
|
|
8DW7
 
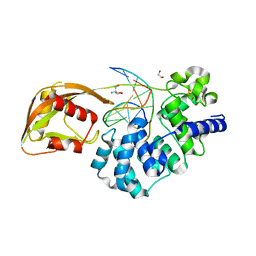 | | DNA glycosylase MutY variant N146S in complex with DNA containing the transition state analog 1N paired with d(8-oxo-G) | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Adenine DNA glycosylase, ... | | Authors: | Demir, M, Russelburg, L.P, Horvath, M.P, David, S.S. | | Deposit date: | 2022-07-31 | | Release date: | 2022-11-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structural snapshots of base excision by the cancer-associated variant MutY N146S reveal a retaining mechanism.
Nucleic Acids Res., 51, 2023
|
|
