8C8W
 
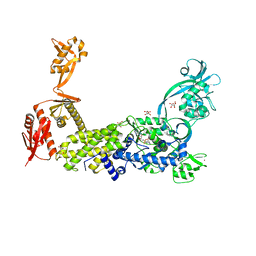 | | Priestia megaterium mupirocin hyper-resistant HIGH motif mutant of type 2 isoleucyl-tRNA synthetase complexed with an isoleucyl-adenylate analogue | | Descriptor: | D(-)-TARTARIC ACID, Isoleucine--tRNA ligase, N-[ISOLEUCINYL]-N'-[ADENOSYL]-DIAMINOSUFONE, ... | | Authors: | Brkic, A, Leibundgut, M, Jablonska, J, Zanki, V, Car, Z, Petrovic Perokovic, V, Ban, N, Gruic-Sovulj, I. | | Deposit date: | 2023-01-21 | | Release date: | 2023-08-16 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Antibiotic hyper-resistance in a class I aminoacyl-tRNA synthetase with altered active site signature motif.
Nat Commun, 14, 2023
|
|
8C9G
 
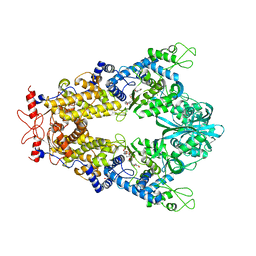 | | Priestia megaterium mupirocin-sensitive isoleucyl-tRNA synthetase 1 complexed with mupirocin | | Descriptor: | CHLORIDE ION, GLYCEROL, Isoleucine--tRNA ligase, ... | | Authors: | Brkic, A, Leibundgut, M, Jablonska, J, Zanki, V, Car, Z, Petrovic Perokovic, V, Ban, N, Gruic-Sovulj, I. | | Deposit date: | 2023-01-22 | | Release date: | 2023-08-23 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Antibiotic hyper-resistance in a class I aminoacyl-tRNA synthetase with altered active site signature motif.
Nat Commun, 14, 2023
|
|
8C9E
 
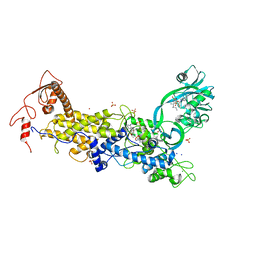 | | Priestia megaterium mupirocin-sensitive isoleucyl-tRNA synthetase 1 complexed with an isoleucyl-adenylate analogue | | Descriptor: | CHLORIDE ION, Isoleucine--tRNA ligase, LITHIUM ION, ... | | Authors: | Brkic, A, Leibundgut, M, Jablonska, J, Zanki, V, Car, Z, Petrovic Perokovic, V, Ban, N, Gruic-Sovulj, I. | | Deposit date: | 2023-01-21 | | Release date: | 2023-08-16 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Antibiotic hyper-resistance in a class I aminoacyl-tRNA synthetase with altered active site signature motif.
Nat Commun, 14, 2023
|
|
1QVG
 
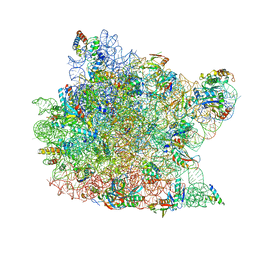 | | Structure of CCA oligonucleotide bound to the tRNA binding sites of the large ribosomal subunit of Haloarcula marismortui | | Descriptor: | 23S ribosomal rna, 50S RIBOSOMAL PROTEIN L10E, 50S ribosomal protein L13P, ... | | Authors: | Schmeing, T.M, Moore, P.B, Steitz, T.A. | | Deposit date: | 2003-08-27 | | Release date: | 2003-11-11 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structures of deacylated tRNA mimics bound to the E site of the large ribosomal subunit
RNA, 9, 2003
|
|
1QVF
 
 | | Structure of a deacylated tRNA minihelix bound to the E site of the large ribosomal subunit of Haloarcula marismortui | | Descriptor: | 23S ribosomal rna, 50S RIBOSOMAL PROTEIN L10E, 50S ribosomal protein L13P, ... | | Authors: | Schmeing, T.M, Moore, P.B, Steitz, T.A. | | Deposit date: | 2003-08-27 | | Release date: | 2003-11-11 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structures of deacylated tRNA mimics bound to the E site of the large ribosomal subunit
RNA, 9, 2003
|
|
6IA2
 
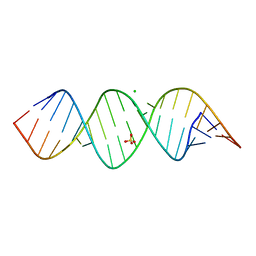 | | Crystal structure of a self-complementary RNA duplex recognized by Com | | Descriptor: | CHLORIDE ION, RNA (5'-R(*AP*GP*AP*GP*AP*AP*CP*CP*CP*GP*GP*AP*GP*UP*UP*CP*CP*CP*U)-3'), SULFATE ION | | Authors: | Nowacka, M, Fernandes, H, Kiliszek, A, Bernat, A, Lach, G, Bujnicki, J.M. | | Deposit date: | 2018-11-26 | | Release date: | 2019-03-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Specific interaction of zinc finger protein Com with RNA and the crystal structure of a self-complementary RNA duplex recognized by Com.
Plos One, 14, 2019
|
|
6V4X
 
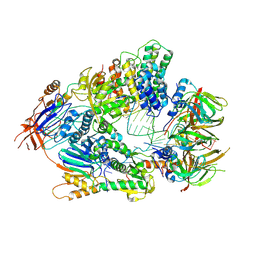 | | Cryo-EM structure of an active human histone pre-mRNA 3'-end processing machinery at 3.2 Angstrom resolution | | Descriptor: | Cleavage and polyadenylation specificity factor subunit 2, Cleavage and polyadenylation specificity factor subunit 3, Small nuclear ribonucleoprotein E, ... | | Authors: | Sun, Y, Zhang, Y, Walz, T, Tong, L. | | Deposit date: | 2019-12-02 | | Release date: | 2020-02-19 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure of an active human histone pre-mRNA 3'-end processing machinery.
Science, 367, 2020
|
|
6O76
 
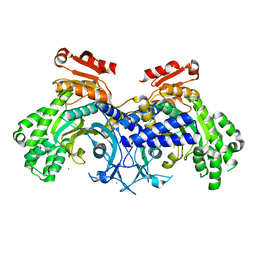 | | Human cytosolic Histidyl-tRNA synthetase (HisRS) with WHEP domain | | Descriptor: | CHLORIDE ION, Histidine--tRNA ligase, cytoplasmic | | Authors: | Kuhle, B, Yang, X.L. | | Deposit date: | 2019-03-07 | | Release date: | 2019-09-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.787 Å) | | Cite: | CMT disease severity correlates with mutation-induced open conformation of histidyl-tRNA synthetase, not aminoacylation loss, in patient cells.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
7UZ0
 
 | | AntiT-tRNA flip UCCA | | Descriptor: | AntiT-tRNA flip UCCA, MAGNESIUM ION, STRONTIUM ION | | Authors: | Price, I.R, Grigg, J.C, Ke, A. | | Deposit date: | 2022-05-07 | | Release date: | 2022-06-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.03 Å) | | Cite: | tRNA Fusion to Streamline RNA Structure Determination: Case Studies in Probing Aminoacyl-tRNA Sensing Mechanisms by the T-Box Riboswitch
Crystals, 12, 2022
|
|
9DRV
 
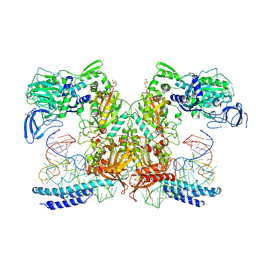 | | Crystal structure of M. tuberculosis PheRS-tRNA complex bound to inhibitor D-004 | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ACETATE ION, ... | | Authors: | Gade, P, Chang, C, Forte, B, Wower, J, Gilbert, I.H, Baragana, B, Michalska, K, Joachimiak, A, Center for Structural Biology of Infectious Diseases (CSBID) | | Deposit date: | 2024-09-26 | | Release date: | 2025-02-12 | | Last modified: | 2025-02-19 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Different chemical scaffolds bind to L-phe site in Mycobacterium tuberculosis Phe-tRNA synthetase.
Eur.J.Med.Chem., 287, 2025
|
|
5J61
 
 | |
5XIP
 
 | | Crystal Structure of Eimeria tenella Prolyl-tRNA Synthetase (EtPRS) in complex with Halofuginone | | Descriptor: | 7-bromo-6-chloro-3-{3-[(2R,3S)-3-hydroxypiperidin-2-yl]-2-oxopropyl}quinazolin-4(3H)-one, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Jain, V, Manickam, Y, Sharma, A. | | Deposit date: | 2017-04-26 | | Release date: | 2018-03-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Targeting Prolyl-tRNA Synthetase to Accelerate Drug Discovery against Malaria, Leishmaniasis, Toxoplasmosis, Cryptosporidiosis, and Coccidiosis
Structure, 25, 2017
|
|
5XIK
 
 | | Crystal Structure of Toxoplasma gondii Prolyl-tRNA Synthetase (TgPRS) in complex with tetrahydro quinazolinone febrifugine | | Descriptor: | 3-[2-oxidanylidene-3-[(2R,3R)-3-oxidanylpiperidin-2-yl]propyl]-5,6,7,8-tetrahydroquinazolin-4-one, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Jain, V, Manickam, Y, Sharma, A. | | Deposit date: | 2017-04-26 | | Release date: | 2018-03-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Targeting Prolyl-tRNA Synthetase to Accelerate Drug Discovery against Malaria, Leishmaniasis, Toxoplasmosis, Cryptosporidiosis, and Coccidiosis
Structure, 25, 2017
|
|
5XIJ
 
 | | Crystal Structure of Toxoplasma gondii Prolyl-tRNA Synthetase (TgPRS) in complex with Inhibitor 9 | | Descriptor: | 3-[(2R)-2-oxidanyl-3-[(2R,3R)-3-oxidanylpiperidin-2-yl]propyl]quinazolin-4-one, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Jain, V, Manickam, Y, Sharma, A. | | Deposit date: | 2017-04-26 | | Release date: | 2018-03-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Targeting Prolyl-tRNA Synthetase to Accelerate Drug Discovery against Malaria, Leishmaniasis, Toxoplasmosis, Cryptosporidiosis, and Coccidiosis
Structure, 25, 2017
|
|
5XIH
 
 | | Crystal Structure of Toxoplasma gondii Prolyl-tRNA Synthetase (TgPRS) in complex with inhibitor 5 | | Descriptor: | 6,7-bis(fluoranyl)-3-[3-[(2R)-3-oxidanylidenepiperidin-2-yl]propyl]quinazolin-4-one, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Jain, V, Manickam, Y, Sharma, A. | | Deposit date: | 2017-04-26 | | Release date: | 2018-03-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Targeting Prolyl-tRNA Synthetase to Accelerate Drug Discovery against Malaria, Leishmaniasis, Toxoplasmosis, Cryptosporidiosis, and Coccidiosis
Structure, 25, 2017
|
|
8C9F
 
 | | Priestia megaterium inactive HIGH motif mutant of type1 isoleucyl-tRNA synthetase complexed with an isoleucyl-adenylate analogue. | | Descriptor: | CHLORIDE ION, Isoleucine--tRNA ligase, LITHIUM ION, ... | | Authors: | Brkic, A, Leibundgut, M, Jablonska, J, Zanki, V, Car, Z, Petrovic Perokovic, V, Ban, N, Gruic-Sovulj, I. | | Deposit date: | 2023-01-21 | | Release date: | 2023-08-16 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Antibiotic hyper-resistance in a class I aminoacyl-tRNA synthetase with altered active site signature motif.
Nat Commun, 14, 2023
|
|
6UGJ
 
 | |
6UGI
 
 | |
8WND
 
 | | Crystal structure of Saccharomyces cerevisiae isoleucyl-tRNA synthetase in complex with tRNA(Ile) and isoleucine | | Descriptor: | 1,2-ETHANEDIOL, ISOLEUCINE, SULFATE ION, ... | | Authors: | Chen, B, Yi, F, Zhou, H. | | Deposit date: | 2023-10-05 | | Release date: | 2024-10-09 | | Last modified: | 2025-01-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The mechanism of discriminative aminoacylation by isoleucyl-tRNA synthetase based on wobble nucleotide recognition.
Nat Commun, 15, 2024
|
|
7K16
 
 | | Tamana Bat Virus xrRNA1 | | Descriptor: | MAGNESIUM ION, RNA (51-MER), SODIUM ION | | Authors: | Jones, R.A, Kieft, J.S. | | Deposit date: | 2020-09-07 | | Release date: | 2020-10-14 | | Last modified: | 2025-05-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Different tertiary interactions create the same important 3D features in a distinct flavivirus xrRNA.
Rna, 27, 2021
|
|
6U9X
 
 | | Structure of T. brucei MERS1-RNA complex | | Descriptor: | Mitochondrial edited mRNA stability factor 1, RNA (5'-R(*GP*AP*GP*AP*GP*GP*GP*GP*GP*UP*U)-3') | | Authors: | Schumacher, M.A. | | Deposit date: | 2019-09-09 | | Release date: | 2019-11-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structures of MERS1, the 5' processing enzyme of mitochondrial mRNAs inTrypanosoma brucei.
Rna, 26, 2020
|
|
6Y3G
 
 | | Crystal structure of phenylalanine tRNA from Escherichia coli | | Descriptor: | CALCIUM ION, GLYCEROL, GUANIDINE, ... | | Authors: | Bourgeois, G, Mechulam, Y, Schmitt, E. | | Deposit date: | 2020-02-18 | | Release date: | 2020-12-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural basis of the interaction between cyclodipeptide synthases and aminoacylated tRNA substrates.
Rna, 26, 2020
|
|
3A6P
 
 | | Crystal structure of Exportin-5:RanGTP:pre-miRNA complex | | Descriptor: | 13-mer peptide, Exportin-5, GTP-binding nuclear protein Ran, ... | | Authors: | Okada, C, Yamashita, E, Lee, S.J, Shibata, S, Katahira, J, Nakagawa, A, Yoneda, Y, Tsukihara, T. | | Deposit date: | 2009-09-07 | | Release date: | 2009-12-08 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.92 Å) | | Cite: | A high-resolution structure of the pre-microRNA nuclear export machinery
Science, 326, 2009
|
|
1GTS
 
 | |
5OMW
 
 | |
