3UQI
 
 | |
3EY6
 
 | | Crystal structure of the FK506-binding domain of human FKBP38 | | Descriptor: | FK506-binding protein 8 | | Authors: | Parthier, C, Maestre-Martinez, M, Neumann, P, Edlich, F, Fischer, G, Luecke, C, Stubbs, M.T. | | Deposit date: | 2008-10-19 | | Release date: | 2009-10-27 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | A charge-sensitive loop in the FKBP38 catalytic domain modulates Bcl-2 binding.
J.Mol.Recognit., 24, 2011
|
|
1YAT
 
 | |
6OQA
 
 | | Crystal structure of CEP250 bound to FKBP12 in the presence of FK506-like novel natural product | | Descriptor: | (3R,4E,7E,10R,11S,12R,13S,16R,17R,24aS)-11,17-dihydroxy-10,12,16-trimethyl-3-[(2R)-1-phenylbutan-2-yl]-6,9,10,11,12,13,14,15,16,17,22,23,24,24a-tetradecahydro-3H-13,17-epoxypyrido[2,1-c][1,4]oxazacyclohenicosine-1,18,19(21H)-trione, 1,2-ETHANEDIOL, 3,6,9,12,15,18,21-HEPTAOXATRICOSANE-1,23-DIOL, ... | | Authors: | Lee, S.-J, Shigdel, U.K, Townson, S.A, Verdine, G.L. | | Deposit date: | 2019-04-26 | | Release date: | 2020-04-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Genomic discovery of an evolutionarily programmed modality for small-molecule targeting of an intractable protein surface.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
2AWG
 
 | | Structure of the PPIase domain of the Human FK506-binding protein 8 | | Descriptor: | 38 kDa FK-506 binding protein | | Authors: | Walker, J.R, Davis, T, Newman, E.M, Finerty, P, Mackenzie, F, Weigelt, J, Sundstrom, M, Arrowsmith, C, Edwards, A, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2005-09-01 | | Release date: | 2005-09-27 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of the human FK-506 binding protein 8
To be Published
|
|
1Y0O
 
 | |
1D6O
 
 | | NATIVE FKBP | | Descriptor: | AMMONIUM ION, PROTEIN (FK506-BINDING PROTEIN), SULFATE ION | | Authors: | Burkhard, P, Taylor, P, Walkinshaw, M.D. | | Deposit date: | 1999-10-15 | | Release date: | 1999-10-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | X-ray structures of small ligand-FKBP complexes provide an estimate for hydrophobic interaction energies.
J.Mol.Biol., 295, 2000
|
|
1D7H
 
 | | FKBP COMPLEXED WITH DMSO | | Descriptor: | AMMONIUM ION, DIMETHYL SULFOXIDE, PROTEIN (FK506-BINDING PROTEIN), ... | | Authors: | Burkhard, P, Taylor, P, Walkinshaw, M.D. | | Deposit date: | 1999-10-18 | | Release date: | 1999-10-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | X-ray structures of small ligand-FKBP complexes provide an estimate for hydrophobic interaction energies.
J.Mol.Biol., 295, 2000
|
|
1D7J
 
 | | FKBP COMPLEXED WITH 4-HYDROXY-2-BUTANONE | | Descriptor: | 4-HYDROXY-2-BUTANONE, AMMONIUM ION, PROTEIN (FK506-BINDING PROTEIN), ... | | Authors: | Burkhard, P, Taylor, P, Walkinshaw, M.D. | | Deposit date: | 1999-10-18 | | Release date: | 1999-10-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | X-ray structures of small ligand-FKBP complexes provide an estimate for hydrophobic interaction energies.
J.Mol.Biol., 295, 2000
|
|
1D7I
 
 | | FKBP COMPLEXED WITH METHYL METHYLSULFINYLMETHYL SULFIDE (DSS) | | Descriptor: | AMMONIUM ION, METHYL METHYLSULFINYLMETHYL SULFIDE, PROTEIN (FK506-BINDING PROTEIN), ... | | Authors: | Burkhard, P, Taylor, P, Walkinshaw, M.D. | | Deposit date: | 1999-10-18 | | Release date: | 1999-10-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | X-ray structures of small ligand-FKBP complexes provide an estimate for hydrophobic interaction energies.
J.Mol.Biol., 295, 2000
|
|
5OBK
 
 | | The Fk1 domain of FKBP51 in complex with (1S,5S,6R)-10-((3,5-dichlorophenyl)sulfonyl)-5-(hydroxymethyl)-3-(pyridin-2-ylmethyl)-3,10-diazabicyclo[4.3.1]decan-2-one | | Descriptor: | (1~{S},5~{S},6~{R})-10-[3,5-bis(chloranyl)phenyl]sulfonyl-5-(hydroxymethyl)-3-(pyridin-2-ylmethyl)-3,10-diazabicyclo[4.3.1]decan-2-one, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Pomplun, S, Sippel, C, Haehle, A, Bracher, A, Hausch, F. | | Deposit date: | 2017-06-28 | | Release date: | 2018-04-04 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Chemogenomic Profiling of Human and Microbial FK506-Binding Proteins.
J. Med. Chem., 61, 2018
|
|
1U79
 
 | | Crystal structure of AtFKBP13 | | Descriptor: | FKBP-type peptidyl-prolyl cis-trans isomerase 3 | | Authors: | Gopalan, G, Swaminathan, K. | | Deposit date: | 2004-08-03 | | Release date: | 2004-09-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural analysis uncovers a role for redox in regulating FKBP13, an immunophilin of the chloroplast thylakoid lumen
Proc.Natl.Acad.Sci.Usa, 101, 2004
|
|
5BXJ
 
 | | Complex of the Fk1 domain mutant A19T of FKBP51 with 4-Nitrophenol | | Descriptor: | P-NITROPHENOL, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Wu, D, Tao, X, Chen, Z, Han, J, Jia, W, Li, X, Wang, Z, He, Y.X. | | Deposit date: | 2015-06-09 | | Release date: | 2016-05-25 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | The environmental endocrine disruptor p-nitrophenol interacts with FKBP51, a positive regulator of androgen receptor and inhibits androgen receptor signaling in human cells
J. Hazard. Mater., 307, 2016
|
|
5DIT
 
 | | The Fk1 domain of FKBP51 in complex with the new synthetic ligand (1R)-3-(3,4-dimethoxyphenyl)-1-f3-[2-(morpholin-4-yl)ethoxy]phenylgpropyl(2S)-1-[(2S,3R)-2-cyclohexyl-3-hydroxybutanoyl]piperidine-2-carboxylate | | Descriptor: | (1R)-3-(3,4-dimethoxyphenyl)-1-{3-[2-(morpholin-4-yl)ethoxy]phenyl}propyl (2S)-1-[(2S,3R)-2-cyclohexyl-3-hydroxybutanoyl]piperidine-2-carboxylate, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Feng, X, Sippel, C, Bracher, A, Hausch, F. | | Deposit date: | 2015-09-01 | | Release date: | 2015-10-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure-Affinity Relationship Analysis of Selective FKBP51 Ligands.
J.Med.Chem., 58, 2015
|
|
5DIV
 
 | | The Fk1 domain of FKBP51 in complex with the new synthetic ligand (S)-N-(1-carbamoylcyclopentyl)-1-((S)-2-cyclohexyl-2-(3,4,5-trimethoxyphenyl)acetyl)piperidine-2-carboxamide | | Descriptor: | (2S)-N-(1-carbamoylcyclopentyl)-1-[(2S)-2-cyclohexyl-2-(3,4,5-trimethoxyphenyl)acetyl]piperidine-2-carboxamide, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Gaali, S, Feng, X, Sippel, C, Bracher, A, Hausch, F. | | Deposit date: | 2015-09-01 | | Release date: | 2016-03-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Rapid, Structure-Based Exploration of Pipecolic Acid Amides as Novel Selective Antagonists of the FK506-Binding Protein 51.
J.Med.Chem., 59, 2016
|
|
1JVW
 
 | | TRYPANOSOMA CRUZI MACROPHAGE INFECTIVITY POTENTIATOR (TCMIP) | | Descriptor: | MACROPHAGE INFECTIVITY POTENTIATOR | | Authors: | Pereira, P.J.B, Vega, M.C, Gonzalez-Rey, E, Fernandez-Carazo, R, Macedo-Ribeiro, S, Gomis-Rueth, F.X, Gonzalez, A, Coll, M. | | Deposit date: | 2001-08-31 | | Release date: | 2002-06-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Trypanosoma cruzi macrophage infectivity potentiator has a rotamase core and a highly exposed alpha-helix.
EMBO Rep., 3, 2002
|
|
3IHZ
 
 | | Crystal structure of the FK506 binding domain of Plasmodium vivax FKBP35 in complex with FK506 | | Descriptor: | 70 kDa peptidylprolyl isomerase, putative, 8-DEETHYL-8-[BUT-3-ENYL]-ASCOMYCIN | | Authors: | Qureshi, I.A, Alag, R, Yoon, H.S, Lescar, J. | | Deposit date: | 2009-07-31 | | Release date: | 2010-06-16 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | NMR and crystallographic structures of the FK506 binding domain of human malarial parasite Plasmodium vivax FKBP35
Protein Sci., 19, 2010
|
|
7OXI
 
 | | ttSlyD with W4A pseudo-wild-type S2 peptide | | Descriptor: | 30S ribosomal protein S2, CHLORIDE ION, Peptidyl-prolyl cis-trans isomerase | | Authors: | Pazicky, S, Lei, J, Loew, C. | | Deposit date: | 2021-06-22 | | Release date: | 2022-03-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Impact of distant peptide substrate residues on enzymatic activity of SlyD.
Cell.Mol.Life Sci., 79, 2022
|
|
7OXK
 
 | | ttSlyD with W4K pseudo-wild-type S2 peptide | | Descriptor: | 30S ribosomal protein S2, Peptidyl-prolyl cis-trans isomerase | | Authors: | Pazicky, S, Lei, J, Loew, C. | | Deposit date: | 2021-06-22 | | Release date: | 2022-03-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Impact of distant peptide substrate residues on enzymatic activity of SlyD.
Cell.Mol.Life Sci., 79, 2022
|
|
7OXG
 
 | | ttSlyD FKBP domain with M8A pseudo-wild-type S2 peptide | | Descriptor: | 30S ribosomal protein S2, CHLORIDE ION, IMIDAZOLE, ... | | Authors: | Pazicky, S, Lei, J, Loew, C. | | Deposit date: | 2021-06-22 | | Release date: | 2022-03-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Impact of distant peptide substrate residues on enzymatic activity of SlyD.
Cell.Mol.Life Sci., 79, 2022
|
|
7OXH
 
 | | ttSlyD with pseudo-wild-type S2 peptide | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, 30S ribosomal protein S2, CHLORIDE ION, ... | | Authors: | Pazicky, S, Lei, J, Loew, C. | | Deposit date: | 2021-06-22 | | Release date: | 2022-03-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Impact of distant peptide substrate residues on enzymatic activity of SlyD.
Cell.Mol.Life Sci., 79, 2022
|
|
7OXJ
 
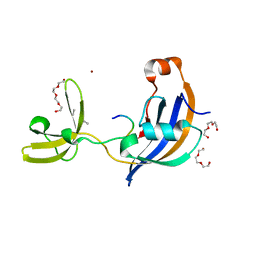 | | ttSlyD with M8A pseudo-wild-type S2 peptide | | Descriptor: | 30S ribosomal protein S2, CHLORIDE ION, Fragment of 30S ribosomal protein S2 peptide, ... | | Authors: | Pazicky, S, Lei, J, Loew, C. | | Deposit date: | 2021-06-22 | | Release date: | 2022-03-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Impact of distant peptide substrate residues on enzymatic activity of SlyD.
Cell.Mol.Life Sci., 79, 2022
|
|
2VN1
 
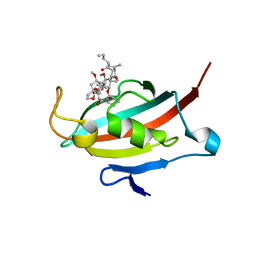 | | Crystal structure of the FK506-binding domain of Plasmodium falciparum FKBP35 in complex with FK506 | | Descriptor: | 70 KDA PEPTIDYLPROLYL ISOMERASE, 8-DEETHYL-8-[BUT-3-ENYL]-ASCOMYCIN | | Authors: | Kotaka, M, Alag, R, Ye, H, Preiser, P.R, Yoon, H.S, Lescar, J. | | Deposit date: | 2008-01-30 | | Release date: | 2008-05-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal Structure of the Fk506 Binding Domain of Plasmodium Falciparum Fkbp35 in Complex with Fk506.
Biochemistry, 47, 2008
|
|
3JYM
 
 | |
3JXV
 
 | |
