6HYT
 
 | | Crystal structure of DHX8 helicase domain bound to ADP at 2.3 Angstrom | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Felisberto-Rodrigues, C, Thomas, J.C, McAndrew, P.C, Le Bihan, Y.V, Burke, R, Workman, P, van Montfort, R.L.M. | | Deposit date: | 2018-10-22 | | Release date: | 2019-08-28 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Structural and functional characterisation of human RNA helicase DHX8 provides insights into the mechanism of RNA-stimulated ADP release.
Biochem.J., 476, 2019
|
|
9B26
 
 | |
7BTK
 
 | | E.coli beta-galactosidase (E537Q) in complex with fluorescent probe KSA01 | | Descriptor: | 4-[[2-[(E)-2-[4-[(2S,3R,4S,5R,6R)-6-(hydroxymethyl)-3,4,5-tris(oxidanyl)oxan-2-yl]oxyphenyl]ethenyl]-3,3-dimethyl-2H-indol-1-yl]methyl]benzoic acid, Beta-galactosidase, DIMETHYL SULFOXIDE, ... | | Authors: | Chen, X, Hu, Y.L, Liu, Q.M, Gao, Y, Yuan, R, Guo, Y. | | Deposit date: | 2020-04-01 | | Release date: | 2021-03-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Two-Dimensional Design Strategy to Construct Smart Fluorescent Probes for the Precise Tracking of Senescence.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
6R2U
 
 | | Zinc-alpha2-Glycoprotein with a Fluorescent Dansyl C 11 Fatty Acid | | Descriptor: | 11-({[5-(dimethylamino)naphthalen-1-yl]sulfonyl}amino)undecanoic acid, AZIDE ION, SULFATE ION, ... | | Authors: | Lau, A.M, Gor, J, Perkins, S.J, Coker, A.R, McDermott, L.C. | | Deposit date: | 2019-03-18 | | Release date: | 2019-09-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Crystal structure of zinc-alpha 2-glycoprotein in complex with a fatty acid reveals multiple different modes of protein-lipid binding.
Biochem.J., 476, 2019
|
|
9AYT
 
 | | Structure of the quorum quenching lactonase GcL bound to N-hexanoyl-L-homoserine lactone | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, COBALT (II) ION, ... | | Authors: | Corbella, M, Bravo, J.A, Demkiv, A.O, Calixto, A.R, Sompiyachoke, K, Bergonzi, C, Kamerlin, S.C.L, Elias, M. | | Deposit date: | 2024-03-08 | | Release date: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Catalytic Redundancies and Conformational Plasticity Drives Selectivity and Promiscuity in Quorum Quenching Lactonases.
Jacs Au, 4, 2024
|
|
6OPH
 
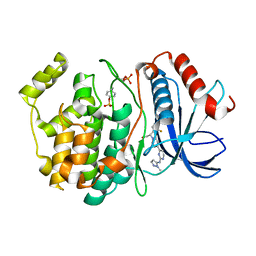 | | phosphorylated ERK2 with GDC-0994 | | Descriptor: | 1-[(1~{S})-1-(4-chloranyl-3-fluoranyl-phenyl)-2-oxidanyl-ethyl]-4-[2-[(2-methylpyrazol-3-yl)amino]pyrimidin-4-yl]pyridin-2-one, Mitogen-activated protein kinase 1 | | Authors: | Vigers, G.P, Smith, D. | | Deposit date: | 2019-04-25 | | Release date: | 2019-07-31 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Activation loop dynamics are controlled by conformation-selective inhibitors of ERK2.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
8PQY
 
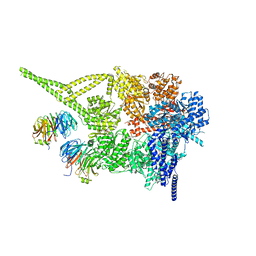 | | Cytoplasmic dynein-1 motor domain bound to LIS1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Cytoplasmic dynein 1 heavy chain 1, ... | | Authors: | Singh, K, Lau, C.K, Manigrasso, G, Gassmann, R, Carter, A.P. | | Deposit date: | 2023-07-12 | | Release date: | 2024-03-27 | | Last modified: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Molecular mechanism of dynein-dynactin complex assembly by LIS1.
Science, 383, 2024
|
|
8J02
 
 | | Human KCNQ2(F104A)-CaM-PIP2-CBD complex in state II | | Descriptor: | Calmodulin-1, Potassium voltage-gated channel subfamily KQT member 2, [(2R)-2-octanoyloxy-3-[oxidanyl-[(1R,2R,3S,4R,5R,6S)-2,3,6-tris(oxidanyl)-4,5-diphosphonooxy-cyclohexyl]oxy-phosphoryl]oxy-propyl] octanoate, ... | | Authors: | Ma, D, Li, X, Guo, J. | | Deposit date: | 2023-04-09 | | Release date: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Ligand activation mechanisms of human KCNQ2 channel.
Nat Commun, 14, 2023
|
|
8XYB
 
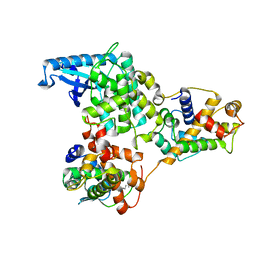 | | hPhK gamma-delta subcomplex in inactive state | | Descriptor: | Calmodulin-1, FARNESYL, Phosphorylase b kinase gamma catalytic chain, ... | | Authors: | Yang, X.K, Xiao, J.Y. | | Deposit date: | 2024-01-19 | | Release date: | 2024-04-03 | | Last modified: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Architecture and activation of human muscle phosphorylase kinase.
Nat Commun, 15, 2024
|
|
8HBE
 
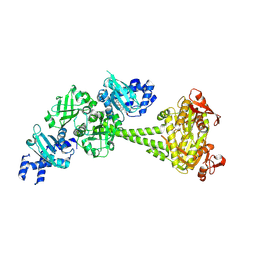 | |
8J01
 
 | | Human KCNQ2-CaM in complex with CBD and PIP2 | | Descriptor: | Calmodulin-1, Potassium voltage-gated channel subfamily KQT member 2, [(2R)-2-octanoyloxy-3-[oxidanyl-[(1R,2R,3S,4R,5R,6S)-2,3,6-tris(oxidanyl)-4,5-diphosphonooxy-cyclohexyl]oxy-phosphoryl]oxy-propyl] octanoate, ... | | Authors: | Ma, D, Li, X, Guo, J. | | Deposit date: | 2023-04-09 | | Release date: | 2023-12-13 | | Last modified: | 2025-09-24 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Ligand activation mechanisms of human KCNQ2 channel.
Nat Commun, 14, 2023
|
|
8XYA
 
 | |
8YHL
 
 | | The Crystal Structure of Tgf-Beta Type I Receptor (Alk5) from Biortus | | Descriptor: | 1,2-ETHANEDIOL, ACETIC ACID, TGF-beta receptor type-1, ... | | Authors: | Wang, F, Cheng, W, Lv, Z, Meng, Q, Xu, Y. | | Deposit date: | 2024-02-28 | | Release date: | 2024-03-13 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | The Crystal Structure of Tgf-Beta Type I Receptor (Alk5) from Biortus
To Be Published
|
|
6I0Z
 
 | | CRYSTAL STRUCTURE OF FASCIN IN COMPLEX WITH COMPOUND 1 | | Descriptor: | Fascin, ~{N}-(2,4-dichlorophenyl)-~{N}-methyl-ethanamide | | Authors: | Schuettelkopf, A.W. | | Deposit date: | 2018-10-27 | | Release date: | 2019-02-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Structure-based design, synthesis and biological evaluation of a novel series of isoquinolone and pyrazolo[4,3-c]pyridine inhibitors of fascin 1 as potential anti-metastatic agents.
Bioorg.Med.Chem.Lett., 29, 2019
|
|
7R1B
 
 | | Mink Variant SARS-CoV-2 Spike with 1 Erect RBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Benton, D.J, Wrobel, A.G, Gamblin, S.J. | | Deposit date: | 2022-02-02 | | Release date: | 2022-03-09 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Evolution of the SARS-CoV-2 spike protein in the human host.
Nat Commun, 13, 2022
|
|
7BDO
 
 | | MAPK14 bound with SR302 | | Descriptor: | Mitogen-activated protein kinase 14, ~{N}-[[4-[[(2~{S})-4-cyclohexyl-1-[[(3~{S})-1-methylsulfonylpiperidin-3-yl]amino]-1-oxidanylidene-butan-2-yl]carbamoyl]phenyl]methyl]imidazo[1,2-a]pyridine-3-carboxamide | | Authors: | Schroeder, M, Roehm, S, Joerger, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-12-22 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Development of a Selective Dual Discoidin Domain Receptor (DDR)/p38 Kinase Chemical Probe.
J.Med.Chem., 64, 2021
|
|
7R16
 
 | | Beta Variant SARS-CoV-2 Spike with 1 Erect RBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Benton, D.J, Wrobel, A.G, Gamblin, S.J. | | Deposit date: | 2022-02-02 | | Release date: | 2022-03-09 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Evolution of the SARS-CoV-2 spike protein in the human host.
Nat Commun, 13, 2022
|
|
6RE4
 
 | | Cryo-EM structure of Polytomella F-ATP synthase, Rotary substate 2B, focussed refinement of F1 head and rotor | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase gamma chain, ... | | Authors: | Murphy, B.J, Klusch, N, Yildiz, O, Kuhlbrandt, W. | | Deposit date: | 2019-04-12 | | Release date: | 2019-07-03 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Rotary substates of mitochondrial ATP synthase reveal the basis of flexible F 1 -F o coupling.
Science, 364, 2019
|
|
7QK4
 
 | | EED in complex with PRC2 allosteric inhibitor compound 22 (MAK683) | | Descriptor: | CHLORIDE ION, Histone-lysine N-methyltransferase EZH2, N-[(5-fluoranyl-2,3-dihydro-1-benzofuran-4-yl)methyl]-8-(2-methylpyridin-3-yl)-[1,2,4]triazolo[4,3-c]pyrimidin-5-amine, ... | | Authors: | Zhao, K, Zhao, M, Luo, X, Zhang, H, Scheufler, C. | | Deposit date: | 2021-12-17 | | Release date: | 2022-04-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.602 Å) | | Cite: | Discovery of the Clinical Candidate MAK683: An EED-Directed, Allosteric, and Selective PRC2 Inhibitor for the Treatment of Advanced Malignancies.
J.Med.Chem., 65, 2022
|
|
8YGW
 
 | | The Crystal Structure of MAPK11 from Biortus | | Descriptor: | 1-(5-TERT-BUTYL-2-P-TOLYL-2H-PYRAZOL-3-YL)-3-[4-(2-MORPHOLIN-4-YL-ETHOXY)-NAPHTHALEN-1-YL]-UREA, Mitogen-activated protein kinase 11 | | Authors: | Wang, F, Cheng, W, Yuan, Z, Lin, D, Guo, S. | | Deposit date: | 2024-02-27 | | Release date: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.301 Å) | | Cite: | The Crystal Structure of MAPK11 from Biortus.
To Be Published
|
|
7QQL
 
 | | The PDZ domain of SNTG2 complexed with the phosphorylated PDZ-binding motif of RSK1 | | Descriptor: | CALCIUM ION, GLYCEROL, Gamma-2-syntrophin,Annexin A2, ... | | Authors: | Cousido-Siah, A, Trave, G, Gogl, G. | | Deposit date: | 2022-01-10 | | Release date: | 2022-04-20 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | A scalable strategy to solve structures of PDZ domains and their complexes.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
6HVJ
 
 | | Human PFKFB3 in complex with a N-Aryl 6-Aminoquinoxaline inhibitor 3 | | Descriptor: | 6-O-phosphono-beta-D-fructofuranose, 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase 3, 8-(3-methyl-1-benzofuran-5-yl)-~{N}-(4-methylsulfonylpyridin-3-yl)quinoxalin-6-amine, ... | | Authors: | Banaszak, K, Pawlik, H, Bialas, A, Fabritius, C.H, Nowak, M. | | Deposit date: | 2018-10-11 | | Release date: | 2018-11-14 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Discovery and Structure-Activity Relationships of N-Aryl 6-Aminoquinoxalines as Potent PFKFB3 Kinase Inhibitors.
ChemMedChem, 14, 2019
|
|
7QJU
 
 | | EED in complex with PRC2 allosteric inhibitor compound 7 | | Descriptor: | CHLORIDE ION, Histone-lysine N-methyltransferase EZH2, N-(2,3-dihydro-1-benzofuran-4-ylmethyl)-8-[4-[(dimethylamino)methyl]phenyl]-[1,2,4]triazolo[4,3-c]pyrimidin-5-amine, ... | | Authors: | Zhao, K, Zhao, M, Luo, X, Zhang, H, Scheufler, C. | | Deposit date: | 2021-12-17 | | Release date: | 2022-04-13 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery of the Clinical Candidate MAK683: An EED-Directed, Allosteric, and Selective PRC2 Inhibitor for the Treatment of Advanced Malignancies.
J.Med.Chem., 65, 2022
|
|
7BRS
 
 | | E.coli beta-galactosidase (E537Q) in complex with fluorescent probe KSA02 | | Descriptor: | 8-[2-[(E)-2-[4-[(2S,3R,4S,5R,6R)-6-(hydroxymethyl)-3,4,5-tris(oxidanyl)oxan-2-yl]oxyphenyl]ethenyl]-3,3-dimethyl-indol-1-ium-1-yl]octanoic acid, Beta-galactosidase, DIMETHYL SULFOXIDE, ... | | Authors: | Chen, X, Hu, Y.L, Gao, Y, Yuan, R, Guo, Y. | | Deposit date: | 2020-03-30 | | Release date: | 2021-03-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Two-Dimensional Design Strategy to Construct Smart Fluorescent Probes for the Precise Tracking of Senescence.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
7QJG
 
 | | EED in complex with PRC2 allosteric inhibitor compound 6 | | Descriptor: | CHLORIDE ION, Histone-lysine N-methyltransferase EZH2, N-(2,3-dihydro-1-benzofuran-7-ylmethyl)-8-[4-[(dimethylamino)methyl]phenyl]-[1,2,4]triazolo[4,3-c]pyrimidin-5-amine, ... | | Authors: | Zhao, K, Zhao, M, Luo, X, Zhang, H, Scheufler, C. | | Deposit date: | 2021-12-16 | | Release date: | 2022-04-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery of the Clinical Candidate MAK683: An EED-Directed, Allosteric, and Selective PRC2 Inhibitor for the Treatment of Advanced Malignancies.
J.Med.Chem., 65, 2022
|
|
