8VPK
 
 | | Structure of Mycobacterium smegmatis 50S ribosomal subunit bound to HflX and erythromycin:50S-HflX-B-Ery | | Descriptor: | 23S ribosomal RNA, 50S Ribosomal Protein L13, 50S Ribosomal Protein L18, ... | | Authors: | Majumdar, S, Koripella, R.K, Sharma, M.R, Manjari, S.R, Banavali, N.K, Agrawal, R.K. | | Deposit date: | 2024-01-16 | | Release date: | 2025-04-30 | | Method: | ELECTRON MICROSCOPY (2.63 Å) | | Cite: | HflX-mediated drug resistance through ribosome splitting and rRNA disordering in mycobacteria.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
7Z64
 
 | | A GH18 from haloalkaliphilic bacterium unveils environment-dependent variations in the catalytic machinery of chitinases | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Chitinase, ... | | Authors: | Madhuprakash, J, Dalhus, B, Rohr, A.K, Bissaro, B, Vaaje-Kolstad, G, Sorlie, M, Eijsink, V.G. | | Deposit date: | 2022-03-10 | | Release date: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | A GH18 from haloalkaliphilic bacterium unveils environment-dependent variations in the catalytic machinery of chitinases
To Be Published
|
|
5W0B
 
 | | Structure of human TUT7 catalytic module (CM) | | Descriptor: | IODIDE ION, SULFATE ION, Terminal uridylyltransferase 7, ... | | Authors: | Faehnle, C.R, Walleshauser, J, Joshua-Tor, L. | | Deposit date: | 2017-05-30 | | Release date: | 2017-06-28 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.614 Å) | | Cite: | Multi-domain utilization by TUT4 and TUT7 in control of let-7 biogenesis.
Nat. Struct. Mol. Biol., 24, 2017
|
|
5ILR
 
 | | H64Q sperm whale myoglobin with a Fe-chlorophenyl moiety | | Descriptor: | (4-chlorophenyl)[3,3'-(7,12-diethenyl-3,8,13,17-tetramethylporphyrin-2,18-diyl-kappa~4~N~21~,N~22~,N~23~,N~24~)di(propanoato)(2-)]iron, GLYCEROL, Myoglobin, ... | | Authors: | Wang, B, Thomas, L.M, Richter-Addo, G.B. | | Deposit date: | 2016-03-04 | | Release date: | 2016-09-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Organometallic myoglobins: Formation of Fe-carbon bonds and distal pocket effects on aryl ligand conformations.
J. Inorg. Biochem., 164, 2016
|
|
8VUF
 
 | | Crystal structure of GH9 (K101P, K103N, V108I) HIV-1 reverse transcriptase in complex with non-nucleoside inhibitor 5i3 | | Descriptor: | (2E)-3-[4-({2-[(1-{[4-(methanesulfonyl)phenyl]methyl}piperidin-4-yl)amino]pyrido[2,3-d]pyrimidin-4-yl}oxy)-3,5-dimethylphenyl]prop-2-enenitrile, 1,2-ETHANEDIOL, DIMETHYL SULFOXIDE, ... | | Authors: | Rumrill, S, Ruiz, F.X, Arnold, E. | | Deposit date: | 2024-01-29 | | Release date: | 2025-05-28 | | Last modified: | 2025-06-11 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Development of enhanced HIV-1 non-nucleoside reverse transcriptase inhibitors with improved resistance and pharmacokinetic profiles.
Sci Adv, 11, 2025
|
|
6VRM
 
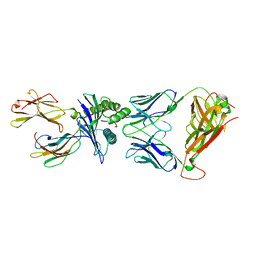 | | T cell receptor-p53-HLA-A2 complex | | Descriptor: | Beta-2-microglobulin, Cellular tumor antigen p53 peptide, MHC class I antigen, ... | | Authors: | Wu, D, Gallagher, D.T, Gowthaman, R, Pierce, B.G, Mariuzza, R.A. | | Deposit date: | 2020-02-08 | | Release date: | 2020-06-17 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Structural basis for oligoclonal T cell recognition of a shared p53 cancer neoantigen.
Nat Commun, 11, 2020
|
|
7MJN
 
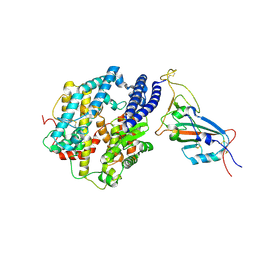 | | Cryo-EM structure of the SARS-CoV-2 N501Y mutant spike protein ectodomain bound to human ACE2 ectodomain (focused refinement of RBD and ACE2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, Spike glycoprotein | | Authors: | Zhu, X, Mannar, D, Srivastava, S.S, Berezuk, A.M, Demers, J.P, Saville, J.W, Leopold, K, Li, W, Dimitrov, D.S, Tuttle, K.S, Zhou, S, Chittori, S, Subramaniam, S. | | Deposit date: | 2021-04-20 | | Release date: | 2021-05-12 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | Cryo-electron microscopy structures of the N501Y SARS-CoV-2 spike protein in complex with ACE2 and 2 potent neutralizing antibodies.
Plos Biol., 19, 2021
|
|
8VYP
 
 | | Cryo-EM Structure of the BRAF V600E monomer | | Descriptor: | 14-3-3 protein zeta/delta, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, Serine/threonine-protein kinase B-raf | | Authors: | Lavoie, H, Lajoie, D, Jin, T, Decossas, M, Maisonneuve, P, Therrien, M. | | Deposit date: | 2024-02-09 | | Release date: | 2025-05-28 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | BRAF oncogenic mutants evade autoinhibition through a common mechanism.
Science, 388, 2025
|
|
6VS8
 
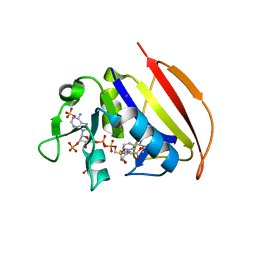 | |
6VSW
 
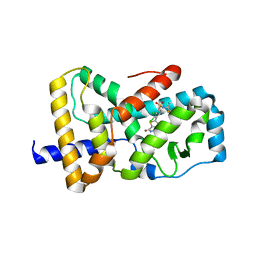 | | Optimization and biological evaluation of thiazole-bis-amide inverse agonists of RORgt | | Descriptor: | 5-(2,3-dichloro-4-{[(2S)-1,1,1-trifluoropropan-2-yl]sulfamoyl}phenyl)-4-(4-fluoropiperidine-1-carbonyl)-N-(2-hydroxy-2-methylpropyl)-1,3-thiazole-2-carboxamide, RAR-related orphan receptor C | | Authors: | Spurlino, J, Milligan, C. | | Deposit date: | 2020-02-12 | | Release date: | 2020-05-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.202 Å) | | Cite: | Optimization and biological evaluation of thiazole-bis-amide inverse agonists of ROR gamma t.
Bioorg.Med.Chem.Lett., 30, 2020
|
|
8VYO
 
 | | Cryo-EM Structure of the BRAF WT monomer | | Descriptor: | 14-3-3 protein zeta/delta, Serine/threonine-protein kinase B-raf, ZINC ION | | Authors: | Lavoie, H, Lajoie, D, Jin, T, Decossas, M, Maisonneuve, P, Therrien, M. | | Deposit date: | 2024-02-09 | | Release date: | 2025-05-28 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.74 Å) | | Cite: | BRAF oncogenic mutants evade autoinhibition through a common mechanism.
Science, 388, 2025
|
|
5ILQ
 
 | | Crystal structure of truncated unliganded Aspartate Transcarbamoylase from Plasmodium falciparum | | Descriptor: | Aspartate carbamoyltransferase, GLYCEROL, SULFATE ION | | Authors: | Lunev, S, Bosch, S.S, Batista, F.D.A, Wrenger, C, Groves, M.R. | | Deposit date: | 2016-03-04 | | Release date: | 2016-06-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of truncated aspartate transcarbamoylase from Plasmodium falciparum.
Acta Crystallogr.,Sect.F, 72, 2016
|
|
5W0Q
 
 | | CREBBP Bromodomain in complex with Cpd17 (N,2,7-trimethyl-2,3-dihydro-4H-benzo[b][1,4]oxazine-4-carboxamide) | | Descriptor: | (2R)-N,2,7-trimethyl-2,3-dihydro-4H-1,4-benzoxazine-4-carboxamide, CREB-binding protein, SULFATE ION | | Authors: | Murray, J.M. | | Deposit date: | 2017-05-31 | | Release date: | 2018-03-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | GNE-781, A Highly Advanced Potent and Selective Bromodomain Inhibitor of Cyclic Adenosine Monophosphate Response Element Binding Protein, Binding Protein (CBP).
J. Med. Chem., 60, 2017
|
|
8VYU
 
 | | Cryo-EM Structure of the BRAF WT monomer bound to PLX8394 | | Descriptor: | (3S)-N-{3-[5-(2-cyclopropylpyrimidin-5-yl)-1H-pyrrolo[2,3-b]pyridine-3-carbonyl]-2,4-difluorophenyl}-3-fluoropyrrolidine-1-sulfonamide, 14-3-3 protein zeta/delta, Serine/threonine-protein kinase B-raf | | Authors: | Lavoie, H, Lajoie, D, Jin, T, Decossas, M, Maisonneuve, P, Therrien, M. | | Deposit date: | 2024-02-09 | | Release date: | 2025-05-28 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (4.07 Å) | | Cite: | BRAF oncogenic mutants evade autoinhibition through a common mechanism.
Science, 388, 2025
|
|
7MMZ
 
 | |
6WBR
 
 | |
8VYW
 
 | | Cryo-EM Structure of the BRAF D594G monomer | | Descriptor: | 14-3-3 protein zeta/delta, Serine/threonine-protein kinase B-raf | | Authors: | Lavoie, H, Lajoie, D, Jin, T, Decossas, M, Maisonneuve, P, Therrien, M. | | Deposit date: | 2024-02-09 | | Release date: | 2025-05-28 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (4.76 Å) | | Cite: | BRAF oncogenic mutants evade autoinhibition through a common mechanism.
Science, 388, 2025
|
|
5VPS
 
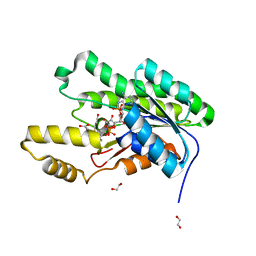 | | Crystal structure of an SDR from Burkholderia ambifaria in complex with NADPH with a TCEP adduct | | Descriptor: | 1,2-ETHANEDIOL, Short-chain dehydrogenase/reductase SDR, [(4~{S})-3-aminocarbonyl-1-[(2~{R},3~{R},4~{S},5~{R})-5-[[[[(2~{R},3~{R},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3-oxidanyl-4-phosphonooxy-oxolan-2-yl]methoxy-oxidanyl-phosphoryl]oxy-oxidanyl-phosphoryl]oxymethyl]-3,4-bis(oxidanyl)oxolan-2-yl]piperidin-4-yl]-tris(3-hydroxy-3-oxopropyl)phosphanium | | Authors: | Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2017-05-05 | | Release date: | 2017-05-24 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Cautionary Tale of Using Tris(alkyl)phosphine Reducing Agents with NAD+-Dependent Enzymes.
Biochemistry, 2020
|
|
6VUH
 
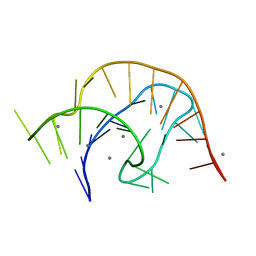 | | APO PreQ1 riboswitch aptamer grown in Mn2+ | | Descriptor: | MANGANESE (II) ION, PREQ1 RIBOSWITCH | | Authors: | Jenkins, J.L, Wedekind, J.E. | | Deposit date: | 2020-02-15 | | Release date: | 2020-06-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.999 Å) | | Cite: | Analysis of a preQ1-I riboswitch in effector-free and bound states reveals a metabolite-programmed nucleobase-stacking spine that controls gene regulation.
Nucleic Acids Res., 48, 2020
|
|
7MHM
 
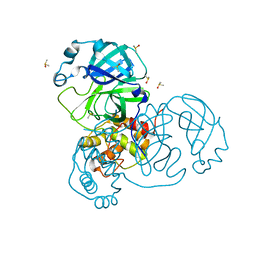 | | Ensemble refinement structure of SARS-CoV-2 main protease (Mpro) at 240 K | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, ZINC ION | | Authors: | Ebrahim, A, Riley, B.T, Kumaran, D, Andi, B, Fuchs, M.R, McSweeney, S, Keedy, D.A. | | Deposit date: | 2021-04-15 | | Release date: | 2021-05-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.5302 Å) | | Cite: | The tem-per-ature-dependent conformational ensemble of SARS-CoV-2 main protease (M pro ).
Iucrj, 9, 2022
|
|
6VUU
 
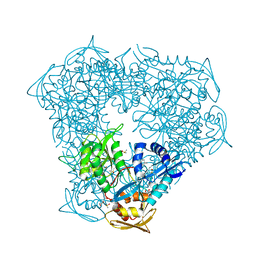 | | Crystal structure of Eis from Mycobacterium tuberculosis in complex with inhibitor SGT1347 | | Descriptor: | (7S)-7-methyl-2-{[3-(piperidin-1-yl)propyl]sulfanyl}-7,8-dihydro[1]benzothieno[2,3-d]pyrimidin-4-amine, DI(HYDROXYETHYL)ETHER, DIMETHYL SULFOXIDE, ... | | Authors: | Punetha, A, Hou, C, Ngo, H.X, Garneau-Tsodikova, S, Tsodikov, O.V. | | Deposit date: | 2020-02-16 | | Release date: | 2020-06-03 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure-Guided Optimization of Inhibitors of Acetyltransferase Eis fromMycobacterium tuberculosis.
Acs Chem.Biol., 15, 2020
|
|
6VIJ
 
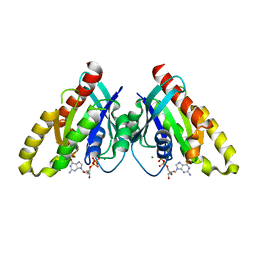 | | Crystal structure of mouse RABL3 in complex with GDP | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Rab-like protein 3 | | Authors: | Su, L, Tomchick, D.R, Beutler, B. | | Deposit date: | 2020-01-13 | | Release date: | 2020-04-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Genetic and structural studies of RABL3 reveal an essential role in lymphoid development and function.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6VIU
 
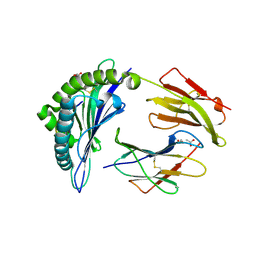 | |
6VV1
 
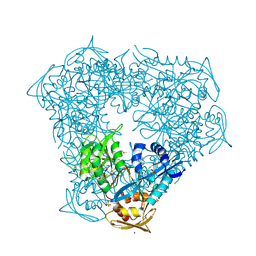 | | Crystal structure of Eis from Mycobacterium tuberculosis in complex with inhibitor SGT384 | | Descriptor: | 2-[(4-amino-6,7-dihydro-5H-cyclopenta[4,5]thieno[2,3-d]pyrimidin-2-yl)sulfanyl]-N-[2-(piperidin-1-yl)ethyl]acetamide, DI(HYDROXYETHYL)ETHER, DIMETHYL SULFOXIDE, ... | | Authors: | Punetha, A, Hou, C, Ngo, H.X, Garneau-Tsodikova, S, Tsodikov, O.V. | | Deposit date: | 2020-02-16 | | Release date: | 2020-06-03 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structure-Guided Optimization of Inhibitors of Acetyltransferase Eis fromMycobacterium tuberculosis.
Acs Chem.Biol., 15, 2020
|
|
5IN5
 
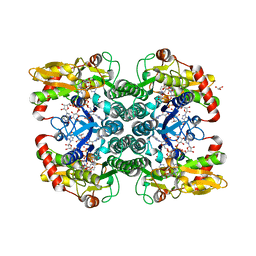 | |
