7ACO
 
 | |
7ACL
 
 | |
7ACM
 
 | |
6B7F
 
 | | Crystal structure of E.coli Phosphopantetheine Adenylyltransferase (PPAT/CoaD) in complex with (R)-3,3-dimethyl-4-(5-vinyl-1H-imidazol-1-yl)isochroman-1-one | | Descriptor: | (4R)-4-(5-ethenyl-1H-imidazol-1-yl)-3,3-dimethyl-3,4-dihydro-1H-2-benzopyran-1-one, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DIMETHYL SULFOXIDE, ... | | Authors: | Proudfoot, A.W, Bussiere, D, Lingel, A. | | Deposit date: | 2017-10-03 | | Release date: | 2017-12-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.562 Å) | | Cite: | High-Confidence Protein-Ligand Complex Modeling by NMR-Guided Docking Enables Early Hit Optimization.
J. Am. Chem. Soc., 139, 2017
|
|
6B7B
 
 | | Crystal structure of E.coli Phosphopantetheine Adenylyltransferase (PPAT/CoaD) in complex with 5-methoxy-2-methyl-1H-indole | | Descriptor: | 5-methoxy-2-methyl-1H-indole, DIMETHYL SULFOXIDE, PYROPHOSPHATE 2-, ... | | Authors: | Proudfoot, A.W, Bussiere, D, Lingel, A. | | Deposit date: | 2017-10-03 | | Release date: | 2017-12-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.981 Å) | | Cite: | High-Confidence Protein-Ligand Complex Modeling by NMR-Guided Docking Enables Early Hit Optimization.
J. Am. Chem. Soc., 139, 2017
|
|
6B7D
 
 | | Crystal structure of E.coli Phosphopantetheine Adenylyltransferase (PPAT/CoaD) in complex with 3-(4-chlorophenyl)-6-methoxy-4,5-dimethylpyridazine | | Descriptor: | 3-(4-chlorophenyl)-6-methoxy-4,5-dimethylpyridazine, DIMETHYL SULFOXIDE, POTASSIUM ION, ... | | Authors: | Proudfoot, A.W, Bussiere, D, Lingel, A. | | Deposit date: | 2017-10-03 | | Release date: | 2017-12-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | High-Confidence Protein-Ligand Complex Modeling by NMR-Guided Docking Enables Early Hit Optimization.
J. Am. Chem. Soc., 139, 2017
|
|
6B7C
 
 | | Crystal structure of E.coli Phosphopantetheine Adenylyltransferase (PPAT/CoaD) in complex with N-((1,3-dimethyl-1H-pyrazol-5-yl)methyl)-5-methyl-1H-imidazo[4,5-b]pyridin-2-amine | | Descriptor: | DI(HYDROXYETHYL)ETHER, DIMETHYL SULFOXIDE, N-[(1,3-dimethyl-1H-pyrazol-5-yl)methyl]-5-methyl-3H-imidazo[4,5-b]pyridin-2-amine, ... | | Authors: | Proudfoot, A.W, Bussiere, D, Lingel, A. | | Deposit date: | 2017-10-03 | | Release date: | 2017-12-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.564 Å) | | Cite: | High-Confidence Protein-Ligand Complex Modeling by NMR-Guided Docking Enables Early Hit Optimization.
J. Am. Chem. Soc., 139, 2017
|
|
3E5O
 
 | | Carbonmonoxy Sperm Whale Myoglobin at 140 K: Laser off | | Descriptor: | CARBON MONOXIDE, Myoglobin, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Tomita, A, Sato, T, Ichiyanagi, K, Nozawa, S, Ichikawa, H, Chollet, M, Kawai, F, Park, S.-Y, Koshihara, S, Adachi, S. | | Deposit date: | 2008-08-14 | | Release date: | 2009-02-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | Visualizing breathing motion of internal cavities in concert with ligand migration in myoglobin.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3E5I
 
 | | Carbonmonoxy Sperm Whale Myoglobin at 120 K: Laser off | | Descriptor: | CARBON MONOXIDE, Myoglobin, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Tomita, A, Sato, T, Ichiyanagi, K, Nozawa, S, Ichikawa, H, Chollet, M, Kawai, F, Park, S.-Y, Koshihara, S, Adachi, S. | | Deposit date: | 2008-08-14 | | Release date: | 2009-02-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | Visualizing breathing motion of internal cavities in concert with ligand migration in myoglobin
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3E4N
 
 | | Carbonmonoxy Sperm Whale Myoglobin at 40 K: Laser off | | Descriptor: | CARBON MONOXIDE, Myoglobin, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Tomita, A, Sato, T, Ichiyanagi, K, Nozawa, S, Ichikawa, H, Chollet, M, Kawai, F, Park, S.-Y, Koshihara, S, Adachi, S. | | Deposit date: | 2008-08-12 | | Release date: | 2009-02-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | Visualizing breathing motion of internal cavities in concert with ligand migration in myoglobin
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3ECZ
 
 | | Carbonmonoxy Sperm Whale Myoglobin at 120 K: Laser on [30 min] | | Descriptor: | CARBON MONOXIDE, Myoglobin, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Tomita, A, Sato, T, Ichiyanagi, K, Nozawa, S, Ichikawa, H, Chollet, M, Kawai, F, Park, S.-Y, Koshihara, S, Adachi, S. | | Deposit date: | 2008-09-02 | | Release date: | 2009-02-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | Visualizing breathing motion of internal cavities in concert with ligand migration in myoglobin
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3EDB
 
 | | Carbonmonoxy Sperm Whale Myoglobin at 120 K: Laser on [150 min] | | Descriptor: | CARBON MONOXIDE, Myoglobin, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Tomita, A, Sato, T, Ichiyanagi, K, Nozawa, S, Ichikawa, H, Chollet, M, Kawai, F, Park, S.-Y, Koshihara, S, Adachi, S. | | Deposit date: | 2008-09-03 | | Release date: | 2009-02-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | Visualizing breathing motion of internal cavities in concert with ligand migration in myoglobin
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3ED9
 
 | | Carbonmonoxy Sperm Whale Myoglobin at 140 K: Laser on [30 min] | | Descriptor: | CARBON MONOXIDE, Myoglobin, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Tomita, A, Sato, T, Ichiyanagi, K, Nozawa, S, Ichikawa, H, Chollet, M, Kawai, F, Park, S.-Y, Koshihara, S, Adachi, S. | | Deposit date: | 2008-09-02 | | Release date: | 2009-02-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | Visualizing breathing motion of internal cavities in concert with ligand migration in myoglobin.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
7T10
 
 | |
8AWI
 
 | | Crystal structure of Human Transthyretin at 1.15 Angstrom resolution | | Descriptor: | SODIUM ION, Transthyretin | | Authors: | Derbyshire, D.J, Hammarstrom, P, von Castelmur, E, Begum, A. | | Deposit date: | 2022-08-29 | | Release date: | 2023-03-01 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Transthyretin Binding Mode Dichotomy of Fluorescent trans -Stilbene Ligands.
Acs Chem Neurosci, 14, 2023
|
|
6ZL2
 
 | |
6ZL9
 
 | |
6ZTE
 
 | |
8OU8
 
 | | Crystal structure of E. coli threonyl tRNA synthetase in complex with a TM84 analogue | | Descriptor: | 1,2-ETHANEDIOL, DIMETHYL SULFOXIDE, Threonine--tRNA ligase, ... | | Authors: | Rodriguez Buitrago, J.A, Parisini, E, Aigars, J. | | Deposit date: | 2023-04-22 | | Release date: | 2023-07-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Synthesis and evaluation of an agrocin 84 toxic moiety (TM84) analogue as a malarial threonyl tRNA synthetase inhibitor.
Org.Biomol.Chem., 21, 2023
|
|
5VPP
 
 | | The 70S P-site tRNA SufA6 complex | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Hong, S, Sunita, S, Dunkle, J.A, Maehigashi, T, Dunham, C.M. | | Deposit date: | 2017-05-05 | | Release date: | 2018-09-26 | | Last modified: | 2018-11-07 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | Mechanism of tRNA-mediated +1 ribosomal frameshifting.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
8FUI
 
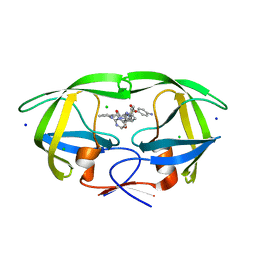 | | HIV-1 wild type protease with GRL-02519A, with N-(2,5-dimethylphenyl)-4-(pyridin-3-yl)pyrimidin-2-amine as P2-P3 group | | Descriptor: | ACETATE ION, CHLORIDE ION, FORMIC ACID, ... | | Authors: | Wang, Y.-F, Wong-Sam, A.E, Ghosh, A.K, Weber, I.T. | | Deposit date: | 2023-01-17 | | Release date: | 2023-05-24 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Exploration of imatinib and nilotinib-derived templates as the P2-Ligand for HIV-1 protease inhibitors: Design, synthesis, protein X-ray structural studies, and biological evaluation.
Eur.J.Med.Chem., 255, 2023
|
|
8A0D
 
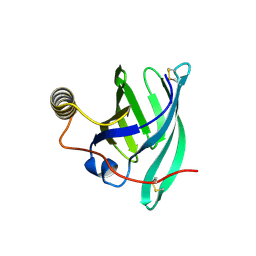 | | Crystal structure of the major guinea pig allergen Cav p 1.0101 part of the lipocalin family | | Descriptor: | Allergen lipocalin Cav p 1 isoform 1 | | Authors: | Herman, R, Charlier, P, Janssen-Weets, B, Hilger, C, Swiontek, K. | | Deposit date: | 2022-05-27 | | Release date: | 2022-08-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.685 Å) | | Cite: | Mammalian derived lipocalin and secretoglobin respiratory allergens strongly bind ligands with potentially immune modulating properties.
Front Allergy, 3, 2022
|
|
3F7D
 
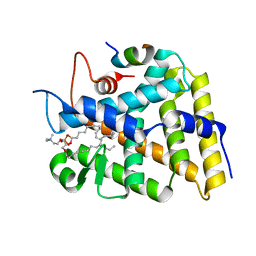 | | SF-1 LBD bound by phosphatidylcholine | | Descriptor: | (2S)-2-{[(1R)-1-hydroxyhexadecyl]oxy}-3-{[(1R)-1-hydroxyoctadecyl]oxy}propyl 2-(trimethylammonio)ethyl phosphate, Peroxisome proliferator-activated receptor gamma coactivator 1-alpha, nuclear receptor SF-1 | | Authors: | Sablin, E.P, Fletterick, R.J. | | Deposit date: | 2008-11-07 | | Release date: | 2008-12-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of SF-1 bound by different phospholipids: evidence for regulatory ligands.
Mol.Endocrinol., 23, 2009
|
|
4NXP
 
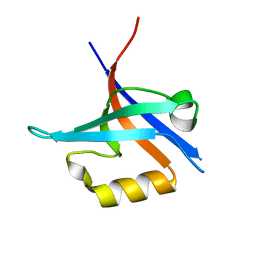 | | Crystal Structure of Free T-cell Lymphoma Invasion and Metastasis-1 PDZ Domain Quadruple Mutant (QM) | | Descriptor: | T-lymphoma invasion and metastasis-inducing protein 1 | | Authors: | Liu, X, Speckhard, D.C, Shepherd, T.R, Hengel, S.R, Fuentes, E.J. | | Deposit date: | 2013-12-09 | | Release date: | 2015-05-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Distinct Roles for Conformational Dynamics in Protein-Ligand Interactions.
Structure, 24, 2016
|
|
3KRQ
 
 | | Crystal structure of the complex of lactoperoxidase with a potent inhibitor amino-triazole at 2.2a resolution | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 3-AMINO-1,2,4-TRIAZOLE, ... | | Authors: | Singh, A.K, Singh, N, Sinha, M, Kushwaha, G.S, Kaur, P, Srinivasan, A, Sharma, S, Singh, T.P. | | Deposit date: | 2009-11-19 | | Release date: | 2010-05-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | First structural evidence for the mode of diffusion of aromatic ligands and ligand-induced closure of the hydrophobic channel in heme peroxidases
J.Biol.Inorg.Chem., 15, 2010
|
|
