8A12
 
 | | Plasmodium falciparum Myosin A full-length, post-rigor state complexed to Mg.ATP-gamma-S | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Moussaoui, D, Robblee, J.P, Auguin, D, Fisher, F, Fagnant, P.M, MacFarlane, J.E, Mueller-Dieckmann, C, Baum, J, Robert-Paganin, J, Trybus, K.M, Houdusse, A. | | Deposit date: | 2022-05-31 | | Release date: | 2023-06-21 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Mechanism of small molecule inhibition of Plasmodium falciparum myosin A informs antimalarial drug design.
Nat Commun, 14, 2023
|
|
7QTB
 
 | | S1 nuclease from Aspergillus oryzae in complex with cytidine-5'-monophosphate | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Adamkova, K, Koval, T, Kolenko, P, Oestergaard, L.H, Dohnalek, J. | | Deposit date: | 2022-01-14 | | Release date: | 2022-11-09 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.04 Å) | | Cite: | Atomic resolution studies of S1 nuclease complexes reveal details of RNA interaction with the enzyme despite multiple lattice-translocation defects.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
8VZG
 
 | | Carbonic anhydrase II mutant with DRD function | | Descriptor: | 5-ACETAMIDO-1,3,4-THIADIAZOLE-2-SULFONAMIDE, CITRATE ANION, Carbonic anhydrase 2, ... | | Authors: | Sethi, D.K. | | Deposit date: | 2024-02-11 | | Release date: | 2024-12-11 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Functional regulation of membrane-bound cytokine activity in engineered T cells via drug-responsive domains derived from carbonic anhydrase 2
To Be Published
|
|
7QTA
 
 | | S1 nuclease from Aspergillus oryzae in complex with uridine | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Nuclease S1, ... | | Authors: | Adamkova, K, Koval, T, Kolenko, P, Oestergaard, L.H, Dohnalek, J. | | Deposit date: | 2022-01-14 | | Release date: | 2022-11-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.06 Å) | | Cite: | Atomic resolution studies of S1 nuclease complexes reveal details of RNA interaction with the enzyme despite multiple lattice-translocation defects.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
6WAE
 
 | | Crystal Structure of 6X-His tagged SmcR | | Descriptor: | 1,2-ETHANEDIOL, LuxR family transcriptional regulator, SULFATE ION | | Authors: | Newman, J.D, Russell, M.M, Gonzalez-Gutierrez, G, van Kessel, J.C. | | Deposit date: | 2020-03-25 | | Release date: | 2020-06-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | The DNA binding domain of the Vibrio vulnificus SmcR transcription factor is flexible and binds diverse DNA sequences.
Nucleic Acids Res., 49, 2021
|
|
7QRC
 
 | | X-ray structure of Trypanosoma cruzi PEX14 in complex with a PEX5-PEX14 PPI inhibitor | | Descriptor: | GLYCEROL, Peroxin-14, ~{N}-(5-ethyl-6-oxidanylidene-benzo[b][1,4]benzothiazepin-2-yl)-2-(4-fluorophenyl)ethanamide | | Authors: | Napolitano, V, Popowicz, G.M, Dawidowski, M, Dubin, G. | | Deposit date: | 2022-01-10 | | Release date: | 2022-11-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Structure-based design, synthesis and evaluation of a novel family of PEX5-PEX14 interaction inhibitors against Trypanosoma.
Eur.J.Med.Chem., 243, 2022
|
|
7NY9
 
 | |
6NJH
 
 | | Crystal Structure of the PDE4D Catalytic Domain and UCR2 Regulatory Helix with T-48 | | Descriptor: | 2-(4-{[4-(3-chlorophenyl)-6-ethyl-1,3,5-triazin-2-yl]amino}phenyl)acetamide, MAGNESIUM ION, ZINC ION, ... | | Authors: | Fox III, D, Fairman, J.W, Gurney, M.E. | | Deposit date: | 2019-01-03 | | Release date: | 2019-05-08 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Design and Synthesis of Selective Phosphodiesterase 4D (PDE4D) Allosteric Inhibitors for the Treatment of Fragile X Syndrome and Other Brain Disorders.
J.Med.Chem., 62, 2019
|
|
5IHE
 
 | | D-family DNA polymerase - DP1 subunit (3'-5' proof-reading exonuclease) | | Descriptor: | 1,2-ETHANEDIOL, 2'-DEOXYADENOSINE-5'-MONOPHOSPHATE, ACETATE ION, ... | | Authors: | Sauguet, L, Raia, P, De Larue, M. | | Deposit date: | 2016-02-29 | | Release date: | 2016-08-31 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Shared active site architecture between archaeal PolD and multi-subunit RNA polymerases revealed by X-ray crystallography.
Nat Commun, 7, 2016
|
|
6D9F
 
 | | Protein 60 with aldehyde deformylating oxidase activity from Kitasatospora setae | | Descriptor: | 1,2-ETHANEDIOL, FE (III) ION, Putative VlmB homolog, ... | | Authors: | Arenas, R, Wilson, D.K, Mak, W.S, Siegel, J.B. | | Deposit date: | 2018-04-28 | | Release date: | 2019-05-08 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Discovery, Design, and Structural Characterization of Alkane-Producing Enzymes across the Ferritin-like Superfamily.
Biochemistry, 59, 2020
|
|
9GQO
 
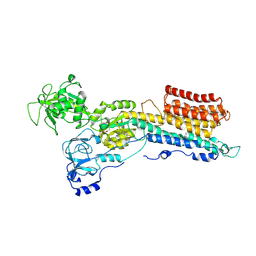 | | Structure of a Ca2+ bound phosphoenzyme intermediate in the inward-to-outward transition of Ca2+-ATPase 1 from Listeria monocytogenes | | Descriptor: | CALCIUM ION, Calcium-transporting ATPase lmo0841, MAGNESIUM ION | | Authors: | Hansen, S.B, Flygaard, R.F, Kjaergaard, M, Nissen, P. | | Deposit date: | 2024-09-09 | | Release date: | 2024-10-16 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.46 Å) | | Cite: | Structure of the [Ca]E2P intermediate of Ca 2+ -ATPase 1 from Listeria monocytogenes.
Embo Rep., 26, 2025
|
|
6NNR
 
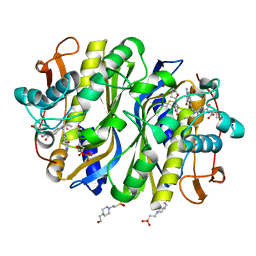 | | high-resolution structure of wild-type E. coli thymidylate synthase | | Descriptor: | 10-PROPARGYL-5,8-DIDEAZAFOLIC ACID, 2'-deoxy-5'-uridylic acid, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Stroud, R.M, Finer-Moore, J. | | Deposit date: | 2019-01-15 | | Release date: | 2019-01-30 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Mg2+ binds to the surface of thymidylate synthase and affects hydride transfer at the interior active site.
J. Am. Chem. Soc., 135, 2013
|
|
8EXA
 
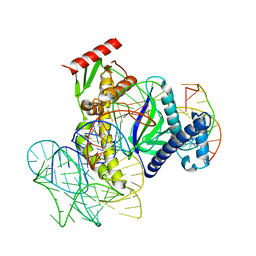 | | ISDra2 TnpB in complex with reRNA and cognate DNA, conformation 1 (RuvC domain resolved) | | Descriptor: | DNA (43-MER), RNA (150-MER), RNA-guided DNA endonuclease TnpB | | Authors: | Sasnauskas, G, Tamulaitiene, G, Carabias, A, Karvelis, T, Druteika, G, Silanskas, A, Montoya, G, Venclovas, C, Kazlauskas, D, Siksnys, V. | | Deposit date: | 2022-10-25 | | Release date: | 2023-04-05 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.14 Å) | | Cite: | TnpB structure reveals minimal functional core of Cas12 nuclease family.
Nature, 616, 2023
|
|
8CDM
 
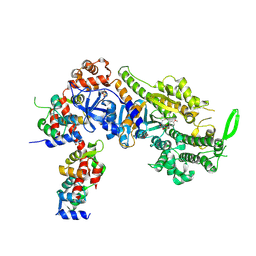 | | Plasmodium falciparum Myosin A full-length, post-rigor state complexed to the inhibitor KNX-002 | | Descriptor: | 1,2-ETHANEDIOL, 1-(4-methoxyphenyl)-~{N}-[(3-thiophen-2-yl-1~{H}-pyrazol-4-yl)methyl]cyclopropan-1-amine, Myosin A tail domain interacting protein, ... | | Authors: | Moussaoui, D, Robblee, J.P, Robert-Paganin, J, Auguin, D, Fisher, F, Fagnant, P.M, MacFarlane, J.E, Mueller-Dieckmann, C, Baum, J, Trybus, K.M, Houdusse, A. | | Deposit date: | 2023-01-31 | | Release date: | 2024-02-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Mechanism of small molecule inhibition of Plasmodium falciparum myosin A informs antimalarial drug design.
Nat Commun, 14, 2023
|
|
7X7G
 
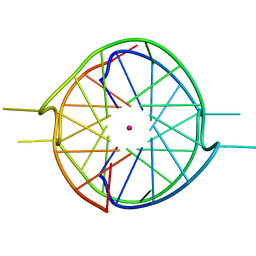 | | Crystal structure of a dimeric interlocked parallel G-quadruplex | | Descriptor: | DNA (5'-D(*GP*GP*GP*GP*TP*GP*GP*GP*AP*GP*GP*TP*GP*GP*GP*T)-3'), POTASSIUM ION | | Authors: | Ngo, K.H, Liew, C.W, Lattmann, S, Winnerdy, F.R, Phan, A.T. | | Deposit date: | 2022-03-09 | | Release date: | 2022-04-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Crystal structures of an HIV-1 integrase aptamer: Formation of a water-mediated A.G.G.G.G pentad in an interlocked G-quadruplex.
Biochem.Biophys.Res.Commun., 613, 2022
|
|
7B52
 
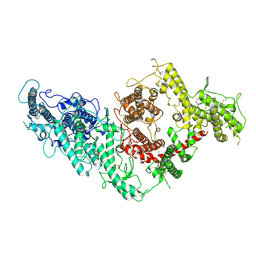 | | VAR2CSA full ectodomain | | Descriptor: | Erythrocyte membrane protein 1 | | Authors: | Wang, K.T, Gourdon, P.E, Dagil, R, Salanti, A. | | Deposit date: | 2020-12-03 | | Release date: | 2021-04-21 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cryo-EM reveals the architecture of placental malaria VAR2CSA and provides molecular insight into chondroitin sulfate binding.
Nat Commun, 12, 2021
|
|
6NCL
 
 | |
6W1B
 
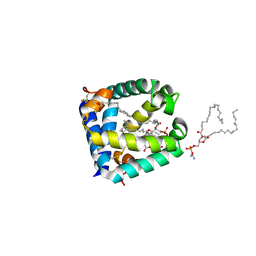 | |
8HCD
 
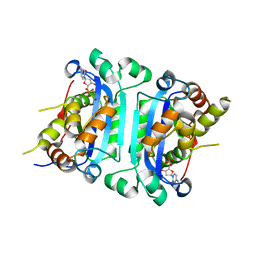 | | Crystal structure of mTREX1-DNA product complex (dNMP) | | Descriptor: | 2'-DEOXYCYTIDINE-5'-MONOPHOSPHATE, 2'-DEOXYGUANOSINE-5'-MONOPHOSPHATE, COBALT (II) ION, ... | | Authors: | Hsiao, Y.Y, Huang, K.W, Wu, C.Y. | | Deposit date: | 2022-11-01 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular insight into the specific enzymatic properties of TREX1 revealing the diverse functions in processing RNA and DNA/RNA hybrids.
Nucleic Acids Res., 51, 2023
|
|
3LEF
 
 | | Crystal structure of HIV epitope-scaffold 4E10_S0_1Z6NA_001 | | Descriptor: | 1,2-ETHANEDIOL, Uncharacterized protein 4E10_S0_1Z6NA_001 (T18) | | Authors: | Holmes, M.A. | | Deposit date: | 2010-01-14 | | Release date: | 2010-09-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Computational Design of Epitope-Scaffolds Allows Induction of Antibodies Specific for a Poorly Immunogenic HIV Vaccine Epitope.
Structure, 18, 2010
|
|
9GB9
 
 | |
8ZFO
 
 | |
8ZFR
 
 | |
8HCH
 
 | | Crystal structure of mTREX1-Uridine complex | | Descriptor: | MAGNESIUM ION, Three-prime repair exonuclease 1, URIDINE | | Authors: | Hsiao, Y.Y, Huang, K.W, Wu, C.Y. | | Deposit date: | 2022-11-01 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular insight into the specific enzymatic properties of TREX1 revealing the diverse functions in processing RNA and DNA/RNA hybrids.
Nucleic Acids Res., 51, 2023
|
|
8ZFP
 
 | |
