8EPR
 
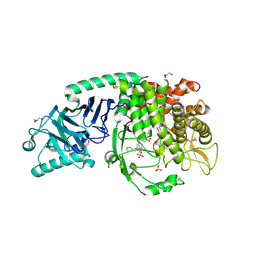 | | Co-crystal structure of Chaetomium glucosidase with compound 19 | | Descriptor: | (2R,3R,4R,5S)-2-(hydroxymethyl)-1-{[3-({[(5M)-3-(methanesulfonyl)-5-(pyridazin-3-yl)phenyl]amino}methyl)phenyl]methyl}piperidine-3,4,5-triol, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Karade, S.S, Mariuzza, R.A. | | Deposit date: | 2022-10-06 | | Release date: | 2023-02-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structure-Based Design of Potent Iminosugar Inhibitors of Endoplasmic Reticulum alpha-Glucosidase I with Anti-SARS-CoV-2 Activity.
J.Med.Chem., 66, 2023
|
|
8CHE
 
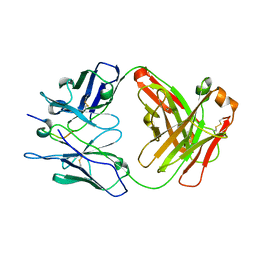 | |
6N5G
 
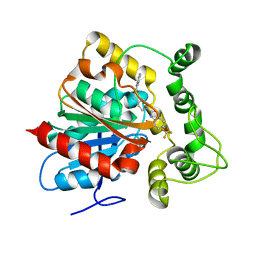 | | Crystal structure of an epoxide hydrolase from Trichoderma reesei in complex with inhibitor 2 | | Descriptor: | 4-[(quinolin-3-yl)methyl]-N-[4-(trifluoromethoxy)phenyl]piperidine-1-carboxamide, Epoxide hydrolase TrEH | | Authors: | Oliveira, G.S, Adriani, P.P, Ribeiro, J.A, Morisseau, C, Hammock, B.D, Dias, M.V, Chambergo, F.S. | | Deposit date: | 2018-11-21 | | Release date: | 2019-11-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The molecular structure of an epoxide hydrolase from Trichoderma reesei in complex with urea or amide-based inhibitors.
Int. J. Biol. Macromol., 129, 2019
|
|
8EQ7
 
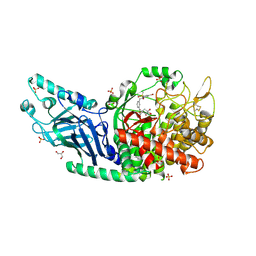 | | Co-crystal structure of Chaetomium glucosidase with compound 20 | | Descriptor: | (2R,3R,4R,5S)-1-[(3-{[3-bromo-5-(methanesulfonyl)anilino]methyl}phenyl)methyl]-2-(hydroxymethyl)piperidine-3,4,5-triol, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Karade, S.S, Mariuzza, R.A. | | Deposit date: | 2022-10-07 | | Release date: | 2023-02-22 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Structure-Based Design of Potent Iminosugar Inhibitors of Endoplasmic Reticulum alpha-Glucosidase I with Anti-SARS-CoV-2 Activity.
J.Med.Chem., 66, 2023
|
|
8ER4
 
 | | Co-crystal structure of Chaetomium glucosidase with compound 23 | | Descriptor: | (2R,3R,4R,5S)-1-(2-{6-[2-(4-azido-2-nitroanilino)ethyl]pyrazin-2-yl}ethyl)-2-(hydroxymethyl)piperidine-3,4,5-triol, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Karade, S.S, Mariuzza, R.A. | | Deposit date: | 2022-10-11 | | Release date: | 2023-02-22 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-Based Design of Potent Iminosugar Inhibitors of Endoplasmic Reticulum alpha-Glucosidase I with Anti-SARS-CoV-2 Activity.
J.Med.Chem., 66, 2023
|
|
8K5R
 
 | | CDK9/cyclin T1 in complex with KB-0742 | | Descriptor: | (1S,3S)-N3-(5-pentan-3-ylpyrazolo[1,5-a]pyrimidin-7-yl)cyclopentane-1,3-diamine, Cyclin-T1, Cyclin-dependent kinase 9 | | Authors: | Zhou, M, Li, H, Gao, H, Trotter, B.W, Freeman, D. | | Deposit date: | 2023-07-24 | | Release date: | 2023-12-06 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.751 Å) | | Cite: | Discovery of KB-0742, a Potent, Selective, Orally Bioavailable Small Molecule Inhibitor of CDK9 for MYC-Dependent Cancers.
J.Med.Chem., 66, 2023
|
|
8K5V
 
 | | Crystal structure of human proMMP-9 catalytic domain in complex with inhibitor | | Descriptor: | 6,7-dihydro-4H-[1,3]oxazolo[4,5-c]pyridin-5-yl-(7-ethyl-2H-indazol-3-yl)methanone, CALCIUM ION, DIHYDROGENPHOSPHATE ION, ... | | Authors: | Kamitani, M, Mima, M, Nishikawa-Shimono, R. | | Deposit date: | 2023-07-24 | | Release date: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Discovery of novel indole derivatives as potent and selective inhibitors of proMMP-9 activation.
Bioorg.Med.Chem.Lett., 97, 2023
|
|
8H8N
 
 | | Crystal structure of apo-R52Y/E56Y/R59Y/E63Y-rHLFr | | Descriptor: | 1,2-ETHANEDIOL, CADMIUM ION, CHLORIDE ION, ... | | Authors: | Hishikawa, Y, Noya, H, Maity, B, Abe, S, Ueno, T. | | Deposit date: | 2022-10-23 | | Release date: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Elucidating Conformational Dynamics and Thermostability of Designed Aromatic Clusters by Using Protein Cages.
Chemistry, 29, 2023
|
|
8H8L
 
 | | Crystal structure of apo-R52F/E56F/R59F/E63F-rHLFr | | Descriptor: | 1,2-ETHANEDIOL, CADMIUM ION, CHLORIDE ION, ... | | Authors: | Hishikawa, Y, Noya, H, Maity, B, Abe, S, Ueno, T. | | Deposit date: | 2022-10-23 | | Release date: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Elucidating Conformational Dynamics and Thermostability of Designed Aromatic Clusters by Using Protein Cages.
Chemistry, 29, 2023
|
|
8H8M
 
 | | Crystal structure of apo-E53F/E57F/E60F/E64F-rHLFr | | Descriptor: | 1,2-ETHANEDIOL, CADMIUM ION, CHLORIDE ION, ... | | Authors: | Hishikawa, Y, Noya, H, Maity, B, Abe, S, Ueno, T. | | Deposit date: | 2022-10-23 | | Release date: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Elucidating Conformational Dynamics and Thermostability of Designed Aromatic Clusters by Using Protein Cages.
Chemistry, 29, 2023
|
|
7OK8
 
 | |
8F16
 
 | | Structure of the STUB1 TPR domain in complex with H203, an all-D Helicon Polypeptide | | Descriptor: | 1,2-ETHANEDIOL, E3 ubiquitin-protein ligase CHIP, N,N'-(1,4-phenylene)diacetamide, ... | | Authors: | Li, K, Callahan, A.J, Travaline, T.L, Tokareva, O.S, Swiecicki, J.-M, Verdine, G.L, Pentelute, B.L, McGee, J.H. | | Deposit date: | 2022-11-04 | | Release date: | 2023-02-15 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Single-Shot Flow Synthesis of D-Proteins for Mirror-Image Phage Display
Chemrxiv, 2023
|
|
5I2O
 
 | | Structure of EF-hand containing protein | | Descriptor: | CALCIUM ION, EF-hand domain-containing protein D2 | | Authors: | Park, K.R, Kwon, M.S, An, J.Y, Lee, J.G, Youn, H.S, Lee, Y, Kang, J.Y, Kim, T.G, Lim, J.J, Park, J.S, Lee, S.H, Song, W.K, Cheong, H, Jun, C, Eom, S.H. | | Deposit date: | 2016-02-09 | | Release date: | 2016-12-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.952 Å) | | Cite: | Structural implications of Ca(2+)-dependent actin-bundling function of human EFhd2/Swiprosin-1.
Sci Rep, 6, 2016
|
|
8F10
 
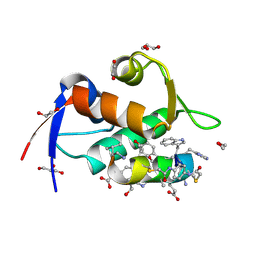 | | Structure of the MDM2 P53 binding domain in complex with H102, an all-D Helicon Polypeptide | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, E3 ubiquitin-protein ligase Mdm2, ... | | Authors: | Li, K, Callahan, A.J, Travaline, T.L, Tokareva, O.S, Swiecicki, J.-M, Verdine, G.L, Pentelute, B.L, McGee, J.H. | | Deposit date: | 2022-11-04 | | Release date: | 2023-02-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Single-Shot Flow Synthesis of D-Proteins for Mirror-Image Phage Display
Chemrxiv, 2023
|
|
6VUU
 
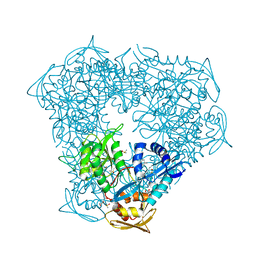 | | Crystal structure of Eis from Mycobacterium tuberculosis in complex with inhibitor SGT1347 | | Descriptor: | (7S)-7-methyl-2-{[3-(piperidin-1-yl)propyl]sulfanyl}-7,8-dihydro[1]benzothieno[2,3-d]pyrimidin-4-amine, DI(HYDROXYETHYL)ETHER, DIMETHYL SULFOXIDE, ... | | Authors: | Punetha, A, Hou, C, Ngo, H.X, Garneau-Tsodikova, S, Tsodikov, O.V. | | Deposit date: | 2020-02-16 | | Release date: | 2020-06-03 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure-Guided Optimization of Inhibitors of Acetyltransferase Eis fromMycobacterium tuberculosis.
Acs Chem.Biol., 15, 2020
|
|
6VV1
 
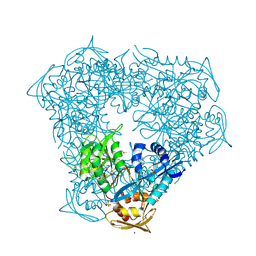 | | Crystal structure of Eis from Mycobacterium tuberculosis in complex with inhibitor SGT384 | | Descriptor: | 2-[(4-amino-6,7-dihydro-5H-cyclopenta[4,5]thieno[2,3-d]pyrimidin-2-yl)sulfanyl]-N-[2-(piperidin-1-yl)ethyl]acetamide, DI(HYDROXYETHYL)ETHER, DIMETHYL SULFOXIDE, ... | | Authors: | Punetha, A, Hou, C, Ngo, H.X, Garneau-Tsodikova, S, Tsodikov, O.V. | | Deposit date: | 2020-02-16 | | Release date: | 2020-06-03 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structure-Guided Optimization of Inhibitors of Acetyltransferase Eis fromMycobacterium tuberculosis.
Acs Chem.Biol., 15, 2020
|
|
6TCR
 
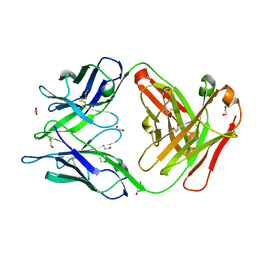 | | Crystal structure of the omalizumab Fab Ser81Arg, Gln83Arg and Leu158Pro light chain mutant | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Omalizumab Fab Ser81Arg, ... | | Authors: | Mitropoulou, A.N, Ceska, T, Beavil, A.J, Henry, A.J, McDonnell, J.M, Sutton, B.J, Davies, A.M. | | Deposit date: | 2019-11-06 | | Release date: | 2020-03-18 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Engineering the Fab fragment of the anti-IgE omalizumab to prevent Fab crystallization and permit IgE-Fc complex crystallization.
Acta Crystallogr.,Sect.F, 76, 2020
|
|
7AVZ
 
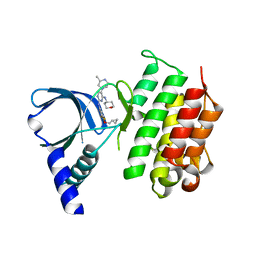 | | MerTK kinase domain in complex with a bisaminopyrimidine inhibitor | | Descriptor: | (R)-N2-(4-(cyclopropylmethoxy)-3,5-difluorophenyl)-5-(3-methylpiperazin-1-yl)-N4-(tetrahydro-2H-pyran-4-yl)pyrimidine-2,4-diamine, Tyrosine-protein kinase Mer | | Authors: | Pflug, A, Nissink, J.W.M, Blackett, C, Goldberg, K, Hennessy, E.J, Hardaker, E, McCoull, W, McMurray, L, Collingwood, O, Overman, R, Preston, M, Rawlins, P, Rivers, E, Schimpl, M, Smith, P, Underwood, E, Truman, C, Warwicker, J, Winter, J, Woodcock, S. | | Deposit date: | 2020-11-06 | | Release date: | 2021-03-03 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Generating Selective Leads for Mer Kinase Inhibitors-Example of a Comprehensive Lead-Generation Strategy.
J.Med.Chem., 64, 2021
|
|
8H8O
 
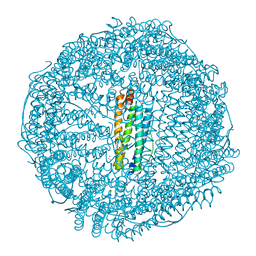 | | Crystal structure of apo-R52W/E56W/R59W/E63W-rHLFr | | Descriptor: | 1,2-ETHANEDIOL, CADMIUM ION, CHLORIDE ION, ... | | Authors: | Hishikawa, Y, Noya, H, Maity, B, Abe, S, Ueno, T. | | Deposit date: | 2022-10-23 | | Release date: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Elucidating Conformational Dynamics and Thermostability of Designed Aromatic Clusters by Using Protein Cages.
Chemistry, 29, 2023
|
|
7AVR
 
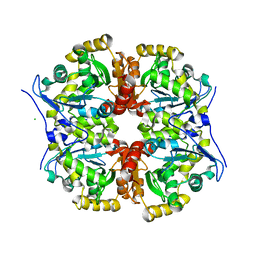 | | The tetrameric structure of haloalkane dehalogenase DpaA from Paraglaciecola agarilytica NO2 | | Descriptor: | CHLORIDE ION, Haloalkane dehalogenase 1 | | Authors: | Mazur, A, Kolenko, P, Prudnikova, T, Grinkevich, P, Kuta Smatanova, I. | | Deposit date: | 2020-11-05 | | Release date: | 2021-03-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The tetrameric structure of the novel haloalkane dehalogenase DpaA from Paraglaciecola agarilytica NO2.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
6NEL
 
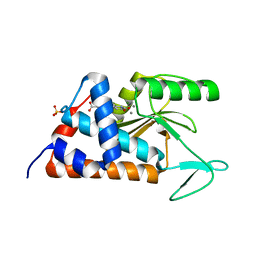 | | 4-(2-(4-fluorophenyl)-5-hydroxy-6-oxo-1,6-dihydropyridin-3-yl)benzoic acid bound to influenza 2009 pH1N1 endonuclease | | Descriptor: | 4-[2-(4-fluorophenyl)-5-hydroxy-6-oxo-1,6-dihydropyridin-3-yl]benzoic acid, MAGNESIUM ION, MANGANESE (II) ION, ... | | Authors: | Bauman, J.D, Arnold, E. | | Deposit date: | 2018-12-17 | | Release date: | 2019-05-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Aryl and Arylalkyl Substituted 3-Hydroxypyridin-2(1H)-ones: Synthesis and Evaluation as Inhibitors of Influenza A Endonuclease.
Chemmedchem, 14, 2019
|
|
6YPG
 
 | |
6Q4V
 
 | | KlenTaq DNA polymerase in complex with dATP | | Descriptor: | 1,2-ETHANEDIOL, 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, DNA (5'-D(*AP*AP*CP*TP*GP*TP*GP*GP*CP*CP*GP*TP*GP*GP*TP*C)-3'), ... | | Authors: | Kropp, H.M, Diederichs, K, Marx, A. | | Deposit date: | 2018-12-06 | | Release date: | 2019-02-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.006 Å) | | Cite: | The Structure of an Archaeal B-Family DNA Polymerase in Complex with a Chemically Modified Nucleotide.
Angew.Chem.Int.Ed.Engl., 58, 2019
|
|
8CDO
 
 | |
8A12
 
 | | Plasmodium falciparum Myosin A full-length, post-rigor state complexed to Mg.ATP-gamma-S | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Moussaoui, D, Robblee, J.P, Auguin, D, Fisher, F, Fagnant, P.M, MacFarlane, J.E, Mueller-Dieckmann, C, Baum, J, Robert-Paganin, J, Trybus, K.M, Houdusse, A. | | Deposit date: | 2022-05-31 | | Release date: | 2023-06-21 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Mechanism of small molecule inhibition of Plasmodium falciparum myosin A informs antimalarial drug design.
Nat Commun, 14, 2023
|
|
