7NQG
 
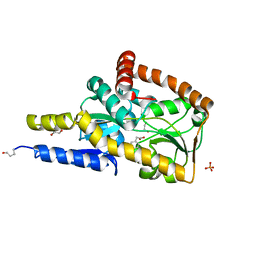 | | The structure of the SBP TarP_Rhp in complex with 4-hydroxyphenylacetate | | Descriptor: | 1,2-ETHANEDIOL, 4-HYDROXYPHENYLACETATE, PHOSPHATE ION, ... | | Authors: | Bisson, C, Salmon, R.C, West, L, Rafferty, J.B, Hitchcock, A, Thomas, G.H, Kelly, D.J. | | Deposit date: | 2021-03-01 | | Release date: | 2021-10-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | The structural basis for high-affinity uptake of lignin-derived aromatic compounds by proteobacterial TRAP transporters.
Febs J., 289, 2022
|
|
7CJD
 
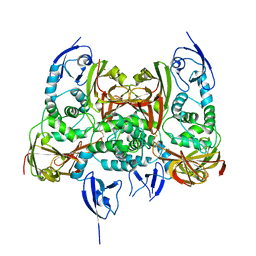 | |
6HMM
 
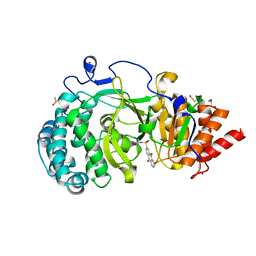 | | POLYADPRIBOSYL GLYCOHYDROLASE IN COMPLEX WITH PDD00013907 | | Descriptor: | DIMETHYL SULFOXIDE, GLYCEROL, Poly(ADP-ribose) glycohydrolase, ... | | Authors: | Tucker, J.A, Brassington, C, Hassall, G. | | Deposit date: | 2018-09-12 | | Release date: | 2018-11-14 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Cell-Active Small Molecule Inhibitors of the DNA-Damage Repair Enzyme Poly(ADP-ribose) Glycohydrolase (PARG): Discovery and Optimization of Orally Bioavailable Quinazolinedione Sulfonamides.
J.Med.Chem., 61, 2018
|
|
8J3J
 
 | | Crystal structure of a bright green fluorescent protein (StayGold) with double mutations (Q140S, Y187F) in jellyfish Cytaeis uchidae from Biortus | | Descriptor: | 1,2-ETHANEDIOL, StayGold(Q140S, Y187F) | | Authors: | Wu, J, Wang, F, Gui, W, Cheng, W, Yang, Y. | | Deposit date: | 2023-04-17 | | Release date: | 2023-12-13 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of a bright green fluorescent protein (StayGold) in jellyfish Cytaeis uchidae from Biortus
To Be Published
|
|
6HQ0
 
 | | Crystal structure of ENL (MLLT1), apo form | | Descriptor: | 1,2-ETHANEDIOL, Protein ENL | | Authors: | Heidenreich, D, Chaikuad, A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-09-22 | | Release date: | 2018-11-28 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Structure-Based Approach toward Identification of Inhibitory Fragments for Eleven-Nineteen-Leukemia Protein (ENL).
J.Med.Chem., 61, 2018
|
|
8AIJ
 
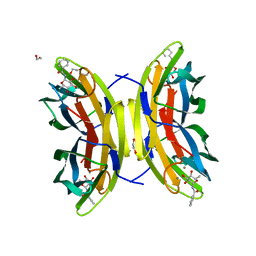 | | STRUCTURE OF THE LECB LECTIN FROM PSEUDOMONAS AERUGINOSA STRAIN PAO1 IN COMPLEX WITH N-(alpha-L-Fucopyranosyl)benzamide (6) | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Fucose-binding lectin PA-IIL, ... | | Authors: | Meiers, J, Mala, P, Varrot, A, Siebs, E, Imberty, A, Titz, A. | | Deposit date: | 2022-07-26 | | Release date: | 2022-11-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Discovery of N -beta-l-Fucosyl Amides as High-Affinity Ligands for the Pseudomonas aeruginosa Lectin LecB.
J.Med.Chem., 65, 2022
|
|
9BLP
 
 | |
9AUA
 
 | | PLP-dependent BesB holoenzyme | | Descriptor: | (3E)-4-{3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}-2-oxobut-3-enoic acid, 1,2-ETHANEDIOL, ACETATE ION, ... | | Authors: | Hedges, J.B, Ryan, K.S. | | Deposit date: | 2024-02-28 | | Release date: | 2025-07-02 | | Last modified: | 2025-07-09 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | Terminal alkyne formation by a pyridoxal phosphate-dependent enzyme.
Nat.Chem.Biol., 2025
|
|
191D
 
 | | CRYSTAL STRUCTURE OF INTERCALATED FOUR-STRANDED D(C3T) | | Descriptor: | DNA (5'-D(*CP*CP*CP*T)-3'), SODIUM ION | | Authors: | Kang, C, Berger, I, Lockshin, C, Ratliff, R, Moyzis, R, Rich, A. | | Deposit date: | 1994-09-29 | | Release date: | 1994-11-30 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of intercalated four-stranded d(C3T) at 1.4 angstroms resolution.
Proc.Natl.Acad.Sci.USA, 91, 1994
|
|
8BS8
 
 | | Bovine naive ultralong antibody AbD08 collected at 100K | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Heavy chain, Light chain | | Authors: | Clarke, J.D, Douangamath, A, Mikolajek, H, Stuart, D.I, Owens, R.J. | | Deposit date: | 2022-11-24 | | Release date: | 2023-05-24 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | The impact of exchanging the light and heavy chains on the structures of bovine ultralong antibodies.
Acta Crystallogr.,Sect.F, 80, 2024
|
|
5D5Z
 
 | | Structure of Chaetomium thermophilum Skn7 coiled-coil domain, crystal form II | | Descriptor: | Putative transcription factor | | Authors: | Neudegger, T, Verghese, J, Hayer-Hartl, M, Bracher, A, Hartl, F.U. | | Deposit date: | 2015-08-11 | | Release date: | 2015-12-30 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of human heat-shock transcription factor 1 in complex with DNA.
Nat.Struct.Mol.Biol., 23, 2016
|
|
9MZT
 
 | |
9MFR
 
 | | Cat DHX9 in Complex with Compound 23 and ADP | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, ... | | Authors: | Lockbaum, G.J, Lee, Y.-T, Sickmier, E.A, Boriack-Sjodin, P.A, Grigoriu, S. | | Deposit date: | 2024-12-10 | | Release date: | 2025-05-07 | | Last modified: | 2025-05-21 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Discovery of ATX968: An Orally Available Allosteric Inhibitor of DHX9.
J.Med.Chem., 68, 2025
|
|
5D2V
 
 | |
7KBX
 
 | |
2K8U
 
 | | Solution NMR structure of trans-4-hydroxynonenal derived dG adduct of (6S,8R,11S)-configuration matched with dC | | Descriptor: | (2S,5R)-5-pentyltetrahydrofuran-2-ol, 5'-D(*DGP*DCP*DTP*DAP*DGP*DCP*DGP*DAP*DGP*DTP*DCP*DC)-3', 5'-D(*DGP*DGP*DAP*DCP*DTP*DCP*DGP*DCP*DTP*DAP*DGP*DC)-3' | | Authors: | Huang, H, Wang, H, Qi, N, Lloyd, R.S, Rizzo, C.J, Stone, M.P. | | Deposit date: | 2008-09-23 | | Release date: | 2008-11-04 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The stereochemistry of trans-4-hydroxynonenal-derived exocyclic 1,N2-2'-deoxyguanosine adducts modulates formation of interstrand cross-links in the 5'-CpG-3' sequence.
Biochemistry, 47, 2008
|
|
7KMU
 
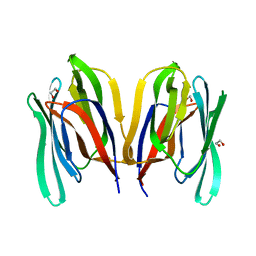 | | Structure of WT Malaysian Banana Lectin | | Descriptor: | 1,2-ETHANEDIOL, Jacalin-type lectin domain-containing protein | | Authors: | Meagher, J.L, Stuckey, J.A. | | Deposit date: | 2020-11-03 | | Release date: | 2021-01-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Targeted disruption of pi-pi stacking in Malaysian banana lectin reduces mitogenicity while preserving antiviral activity.
Sci Rep, 11, 2021
|
|
2HOP
 
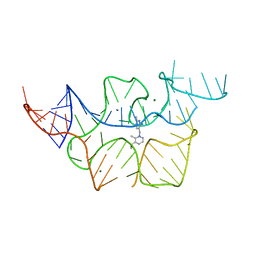 | |
2K76
 
 | | Solution structure of a paralog-specific Mena binder by NMR | | Descriptor: | pGolemi | | Authors: | Link, N.M, Hunke, C, Eichler, J, Bayer, P. | | Deposit date: | 2008-08-03 | | Release date: | 2009-06-16 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The solution structure of pGolemi, a high affinity Mena EVH1 binding miniature protein, suggests explanations for paralog-specific binding to Ena/VASP homology (EVH) 1 domains.
Biol.Chem., 390, 2009
|
|
2K8T
 
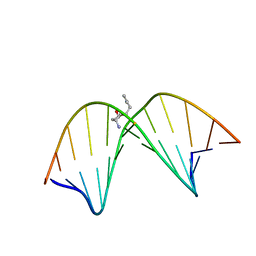 | | Solution NMR structure of trans-4-hydroxynonenal derived dG adduct of (6R,8S,11R)-configuration opposite dC | | Descriptor: | (2S,5R)-5-pentyltetrahydrofuran-2-ol, 5'-D(*DGP*DCP*DTP*DAP*DGP*DCP*DGP*DAP*DGP*DTP*DCP*DC)-3', 5'-D(*DGP*DGP*DAP*DCP*DTP*DCP*DGP*DCP*DTP*DAP*DGP*DC)-3' | | Authors: | Huang, H, Wang, H, Qi, N, Lloyd, R.S, Rizzo, C.J, Stone, M.P. | | Deposit date: | 2008-09-23 | | Release date: | 2008-11-04 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The stereochemistry of trans-4-hydroxynonenal-derived exocyclic 1,N2-2'-deoxyguanosine adducts modulates formation of interstrand cross-links in the 5'-CpG-3' sequence.
Biochemistry, 47, 2008
|
|
7C4D
 
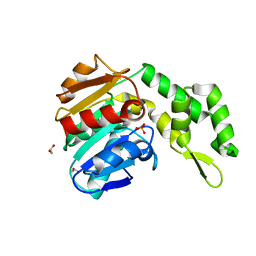 | | Marine microorganism esterase | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Putative esterase, ... | | Authors: | Zhu, C.H, Wu, Y.K, Isupov, M.N. | | Deposit date: | 2020-05-16 | | Release date: | 2021-05-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Structural Insights into a Novel Esterase from the East Pacific Rise and Its Improved Thermostability by a Semirational Design.
J.Agric.Food Chem., 69, 2021
|
|
6UYA
 
 | | Crystal structure of Compound 19 bound to IRAK4 | | Descriptor: | Interleukin-1 receptor-associated kinase 4, N-{2-[(2R)-2-fluoro-3-hydroxy-3-methylbutyl]-6-(morpholin-4-yl)-1-oxo-2,3-dihydro-1H-isoindol-5-yl}pyrazolo[1,5-a]pyrimidine-3-carboxamide, SULFATE ION | | Authors: | Kiefer, J.R, Bryan, M.C, Lupardus, P.J, Zarrin, A.A, Rajapaksa, N.S, Gobbi, A, Drobnick, J, Kolesnikov, A, Liang, J, Do, S. | | Deposit date: | 2019-11-12 | | Release date: | 2019-11-20 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Discovery of Potent Benzolactam IRAK4 Inhibitors with Robust in Vivo Activity.
Acs Med.Chem.Lett., 11, 2020
|
|
7PZU
 
 | | LpxC Inhibitors With Fluoroproline As A Novel Zinc-Binding Group Can Serve As A Novel Class of Antibiotic With Activity Against Multidrug-Resistant Gram-Negative Bacteria | | Descriptor: | (2S,4S)-N-((3R,5R)-1-(cyclopropanecarbonyl)-5-((3-fluoro-4-((4-(morpholinomethyl)phenyl)ethynyl)benzamido)methyl)pyrrolidin-3-yl)-4-fluoropyrrolidine-2-carboxamide, UDP-3-O-acyl-N-acetylglucosamine deacetylase, ZINC ION | | Authors: | Ryan, M.D, Pallin, T.D, Lamers, M.B.A.C, Leonard, P.M. | | Deposit date: | 2021-10-13 | | Release date: | 2022-11-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | LpxC Inhibitors With Fluoroproline As A Novel Zinc-Binding Group Can Serve As A Novel Class of Antibiotic With Activity Against Multidrug-Resistant Gram-Negative Bacteria
To Be Published
|
|
6V0P
 
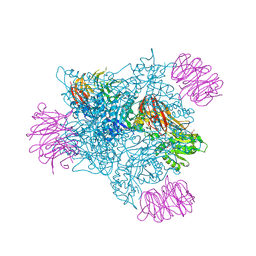 | | PRMT5 complex bound to covalent PBM inhibitor BRD6711 | | Descriptor: | 2-(5-chloro-6-oxopyridazin-1(6H)-yl)-N-(4-methyl-3-sulfamoylphenyl)acetamide, CHLORIDE ION, GLYCEROL, ... | | Authors: | McMillan, B.J, McKinney, D.C. | | Deposit date: | 2019-11-19 | | Release date: | 2020-11-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Discovery of a First-in-Class Inhibitor of the PRMT5-Substrate Adaptor Interaction.
J.Med.Chem., 64, 2021
|
|
8Y7L
 
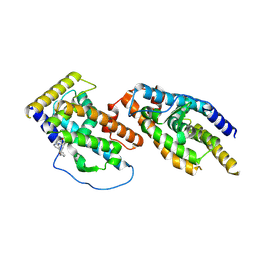 | |
