5W90
 
 | | FEZ-1 metallo-beta-lactamase from Legionella gormanii modelled with unknown ligand | | Descriptor: | FEZ-1 protein, GLYCEROL, SULFATE ION, ... | | Authors: | Garcia-Saez, I, Mercuri, P.S, Kahn, R, Shabalin, I.G, Raczynska, J.E, Jaskolski, M, Minor, W, Papamicael, C, Frere, J.M, Galleni, M, Dideberg, O. | | Deposit date: | 2017-06-22 | | Release date: | 2018-06-20 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Three-dimensional structure of FEZ-1, a monomeric subclass B3 metallo-beta-lactamase from Fluoribacter gormanii, in native form and in complex with D-captopril.
J. Mol. Biol., 325, 2003
|
|
6MOM
 
 | | Crystal structure of human Interleukin-1 receptor associated Kinase 4 (IRAK 4, CID 100300) in complex with compound NCC00371481 (BSI 107591) | | Descriptor: | 1,2-ETHANEDIOL, 6-[7-methoxy-6-(1-methyl-1H-pyrazol-4-yl)imidazo[1,2-a]pyridin-3-yl]-N-[(3R)-pyrrolidin-3-yl]pyridin-2-amine, Interleukin-1 receptor-associated kinase 4 | | Authors: | Abendroth, J, Mayclin, S.J, Lorimer, D.D, Starczynowski, D, Hoyt, S, Tawa, G, Thomas, C. | | Deposit date: | 2018-10-04 | | Release date: | 2019-10-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Overcoming adaptive therapy resistance in AML by targeting immune response pathways.
Sci Transl Med, 11, 2019
|
|
6VKQ
 
 | | Crystal Structure of human PARP-1 CAT domain bound to inhibitor EB-47 | | Descriptor: | 2-[4-[(2S,3S,4R,5R)-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]carbonylpiperazin-1-yl]-N-(1-oxidanylidene-2,3-dihydroisoindol-4-yl)ethanamide, Poly [ADP-ribose] polymerase 1, SULFATE ION | | Authors: | Steffen, J.D, Pascal, J.M. | | Deposit date: | 2020-01-21 | | Release date: | 2020-06-17 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis for allosteric PARP-1 retention on DNA breaks.
Science, 368, 2020
|
|
7QUE
 
 | | The STK17A (DRAK1) Kinase Domain Bound to CKJB68 | | Descriptor: | Serine/threonine-protein kinase 17A, ~{N}-(phenylmethyl)-7,10-dioxa-13,17,18,21-tetrazatetracyclo[12.5.2.1^{2,6}.0^{17,20}]docosa-1(20),2(22),3,5,14(21),15,18-heptaene-5-carboxamide | | Authors: | Mathea, S, Preuss, F, Chatterjee, D, Dederer, V, Kurz, C.G, Amrhein, J.A, Hanke, T, Knapp, S. | | Deposit date: | 2022-01-17 | | Release date: | 2022-02-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Illuminating the Dark: Highly Selective Inhibition of Serine/Threonine Kinase 17A with Pyrazolo[1,5- a ]pyrimidine-Based Macrocycles.
J.Med.Chem., 65, 2022
|
|
7QUF
 
 | | The STK17A (DRAK1) Kinase Domain Bound to CK156 | | Descriptor: | Serine/threonine-protein kinase 17A, ~{N}-~{tert}-butyl-7,10-dioxa-13,17,18,21-tetrazatetracyclo[12.5.2.1^{2,6}.0^{17,20}]docosa-1(20),2(22),3,5,14(21),15,18-heptaene-5-carboxamide | | Authors: | Mathea, S, Preuss, F, Chatterjee, D, Dederer, V, Kurz, C, Hanke, T, Knapp, S. | | Deposit date: | 2022-01-17 | | Release date: | 2022-02-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Illuminating the Dark: Highly Selective Inhibition of Serine/Threonine Kinase 17A with Pyrazolo[1,5- a ]pyrimidine-Based Macrocycles.
J.Med.Chem., 65, 2022
|
|
5LQN
 
 | |
8TNH
 
 | | Cryo-EM structure of HIV-1 Env BG505 DS-SOSIP in complex with broadly neutralizing llama nanobody G36 targeting the CD4-binding site | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CD4-binding site nanobody G36, ... | | Authors: | Zhou, T, Kwong, P.D, Xu, J. | | Deposit date: | 2023-08-01 | | Release date: | 2024-05-08 | | Last modified: | 2024-12-18 | | Method: | ELECTRON MICROSCOPY (3.32 Å) | | Cite: | Ultrapotent Broadly Neutralizing Human-llama Bispecific Antibodies against HIV-1.
Adv Sci, 11, 2024
|
|
8TNI
 
 | | Cryo-EM structure of HIV-1 Env BG505 DS-SOSIP in complex with broadly neutralizing bi-specific antibody CAP256L-R27 targeting the CD4-binding site and the V2-apex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, HIV-1 BG505 DS-SOSIP gp120, ... | | Authors: | Zhou, T, Morano, N.C, Roark, R.S, Kwong, P.D, Xu, J. | | Deposit date: | 2023-08-01 | | Release date: | 2024-05-08 | | Last modified: | 2024-12-18 | | Method: | ELECTRON MICROSCOPY (3.61 Å) | | Cite: | Ultrapotent Broadly Neutralizing Human-llama Bispecific Antibodies against HIV-1.
Adv Sci, 11, 2024
|
|
8TNG
 
 | | Cryo-EM structure of HIV-1 Env BG505 DS-SOSIP in complex with broadly neutralizing llama nanobody R27 targeting the CD4-binding site | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CD4-binding site targeting nanobody R27, ... | | Authors: | Zhou, T, Kwong, P.D, Xu, J. | | Deposit date: | 2023-08-01 | | Release date: | 2024-05-08 | | Last modified: | 2024-12-18 | | Method: | ELECTRON MICROSCOPY (3.58 Å) | | Cite: | Ultrapotent Broadly Neutralizing Human-llama Bispecific Antibodies against HIV-1.
Adv Sci, 11, 2024
|
|
6PXF
 
 | | Structure of human Cathepsin K with an ectosteric inhibitor at 1.85 Angstrom resolution | | Descriptor: | 1,6,6-trimethyl-10,11-dioxo-6,7,8,9,10,11-hexahydrophenanthro[1,2-b]furan-2-sulfonic acid, Cathepsin K | | Authors: | Law, S, Aguda, A.H, Nguyen, N.T, Brayer, G.D, Bromme, D. | | Deposit date: | 2019-07-25 | | Release date: | 2020-07-29 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure of human Cathepsin K with an ectosteric inhibitor at 1.85 Angstrom resolution
To Be Published
|
|
6GSS
 
 | | HUMAN GLUTATHIONE S-TRANSFERASE P1-1, COMPLEX WITH GLUTATHIONE | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLUTATHIONE, GLUTATHIONE S-TRANSFERASE P1-1 | | Authors: | Oakley, A, Parker, M. | | Deposit date: | 1997-08-13 | | Release date: | 1998-09-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The structures of human glutathione transferase P1-1 in complex with glutathione and various inhibitors at high resolution.
J.Mol.Biol., 274, 1997
|
|
5WJJ
 
 | | Structure-based Design, Synthesis, and Biological Evaluation of Imidazo[1,2-b]pyridazine-based p38 MAP Kinase Inhibitors | | Descriptor: | Mitogen-activated protein kinase 14, N-{4-[2-(4-fluoro-3-methylphenyl)imidazo[1,2-b]pyridazin-3-yl]pyridin-2-yl}-2-methyl-1-oxo-1lambda~5~-pyridine-4-carboxamide | | Authors: | Snell, G.P, Okada, K, Bragstad, K, Sang, B.-C. | | Deposit date: | 2017-07-23 | | Release date: | 2018-01-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure-based design, synthesis, and biological evaluation of imidazo[1,2-b]pyridazine-based p38 MAP kinase inhibitors.
Bioorg. Med. Chem., 26, 2018
|
|
7AVQ
 
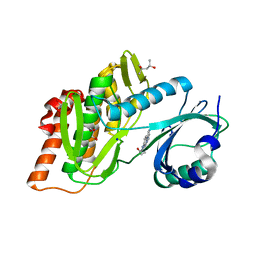 | | Crystal structure of haspin in complex with disubstituted imidazo[1,2- b]pyridazine inhibitor (compound 12) | | Descriptor: | (2~{R})-2-[[3-(2~{H}-indazol-5-yl)imidazo[1,2-b]pyridazin-6-yl]amino]butan-1-ol, (4S)-2-METHYL-2,4-PENTANEDIOL, GLYCEROL, ... | | Authors: | Chaikuad, A, Bonnet, P, Routier, S, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-11-05 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Design of new disubstituted imidazo[1,2- b ]pyridazine derivatives as selective Haspin inhibitors. Synthesis, binding mode and anticancer biological evaluation.
J Enzyme Inhib Med Chem, 35, 2020
|
|
9FKY
 
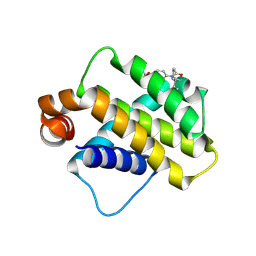 | | Discovery of a Series of Covalent, Cell Active Bfl-1 Inhibitors | | Descriptor: | Bcl-2-related protein A1, ~{N}-[4-[(1~{R},3~{R})-3-azanylcyclopentyl]oxyphenyl]-~{N}-[(1~{S})-1-[3-cyano-4-(trifluoromethyl)phenyl]ethyl]propanamide | | Authors: | Hargreaves, D. | | Deposit date: | 2024-06-04 | | Release date: | 2024-10-02 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.557 Å) | | Cite: | Structure-Based Optimization of a Series of Covalent, Cell Active Bfl-1 Inhibitors.
J.Med.Chem., 67, 2024
|
|
9GCI
 
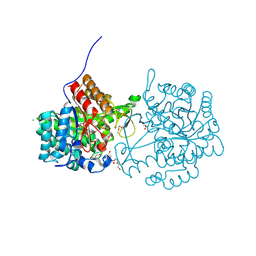 | |
7LE8
 
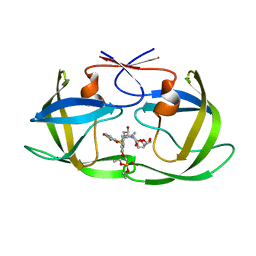 | | HIV-1 Protease WT (NL4-3) in Complex with PD4 (LR4-23) | | Descriptor: | Protease, diethyl [(4-{(2S,3R)-4-{[(2H-1,3-benzodioxol-5-yl)sulfonyl](2-methylpropyl)amino}-2-[({[(3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl]oxy}carbonyl)amino]-3-hydroxybutyl}phenoxy)methyl]phosphonate | | Authors: | Lockbaum, G.J, Rusere, L.N, Henes, M, Kosovrasti, K, Lee, S.K, Spielvogel, E, Nalivaika, E.A, Swanstrom, R, KurtYilmaz, N, Schiffer, C.A, Ali, A. | | Deposit date: | 2021-01-14 | | Release date: | 2022-01-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.644 Å) | | Cite: | HIV-1 Protease Inhibitors with a P1 Phosphonate Modification Maintain Potency against Drug Resistant Variants by Increased van der Waals Contacts with Flaps Residues
To Be Published
|
|
7LEE
 
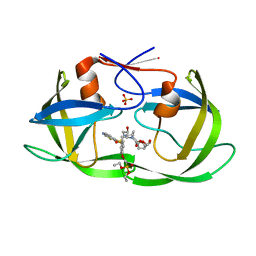 | | HIV-1 Protease WT (NL4-3) in Complex with PU5 (LR4-47) | | Descriptor: | Protease, SULFATE ION, diethyl [(4-{(2S,3R)-4-{[(1,3-benzothiazol-6-yl)sulfonyl][(2S)-2-methylbutyl]amino}-2-[({[(3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl]oxy}carbonyl)amino]-3-hydroxybutyl}phenoxy)methyl]phosphonate | | Authors: | Lockbaum, G.J, Rusere, L.N, Henes, M, Kosovrasti, K, Lee, S.K, Spielvogel, E, Nalivaika, E.A, Swanstrom, R, KurtYilmaz, N, Schiffer, C.A, Ali, A. | | Deposit date: | 2021-01-14 | | Release date: | 2022-01-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.795 Å) | | Cite: | HIV-1 Protease Inhibitors with a P1 Phosphonate Modification Maintain Potency against Drug Resistant Variants by Increased van der Waals Contacts with Flaps Residues
To Be Published
|
|
7LE9
 
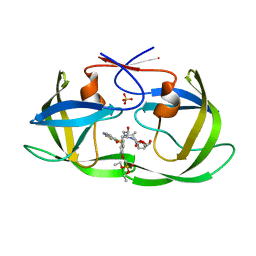 | | HIV-1 Protease WT (NL4-3) in Complex with PD5 (LR4-22) | | Descriptor: | Protease, SULFATE ION, diethyl [(4-{(2S,3R)-4-{[(1,3-benzothiazol-6-yl)sulfonyl](2-methylpropyl)amino}-2-[({[(3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl]oxy}carbonyl)amino]-3-hydroxybutyl}phenoxy)methyl]phosphonate | | Authors: | Lockbaum, G.J, Rusere, L.N, Henes, M, Kosovrasti, K, Lee, S.K, Spielvogel, E, Nalivaika, E.A, Swanstrom, R, KurtYilmaz, N, Schiffer, C.A, Ali, A. | | Deposit date: | 2021-01-14 | | Release date: | 2022-01-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.797 Å) | | Cite: | HIV-1 Protease Inhibitors with a P1 Phosphonate Modification Maintain Potency against Drug Resistant Variants by Increased van der Waals Contacts with Flaps Residues
To Be Published
|
|
7LE4
 
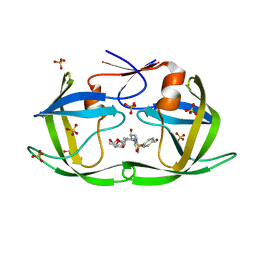 | | HIV-1 Protease WT (NL4-3) in Complex with UMass7 | | Descriptor: | (3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl [(1S,2R)-1-benzyl-3-{(2-ethylbutyl)[(4-methoxyphenyl)sulfonyl]amino}-2-hydroxypropyl]carbamate, Protease, SULFATE ION | | Authors: | Lockbaum, G.J, Rusere, L.N, Henes, M, Kosovrasti, K, Lee, S.K, Spielvogel, E, Nalivaika, E.A, Swanstrom, R, KurtYilmaz, N, Schiffer, C.A, Ali, A. | | Deposit date: | 2021-01-14 | | Release date: | 2022-01-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.993 Å) | | Cite: | HIV-1 Protease Inhibitors with a P1 Phosphonate Modification Maintain Potency against Drug Resistant Variants by Increased van der Waals Contacts with Flaps Residues
To Be Published
|
|
7LEI
 
 | | HIV-1 Protease WT (NL4-3) in Complex with PU10 (LR4-07) | | Descriptor: | Protease, SULFATE ION, diethyl [(4-{(2S,3R)-4-{[(1,3-benzothiazol-6-yl)sulfonyl](2-ethylbutyl)amino}-2-[({[(3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl]oxy}carbonyl)amino]-3-hydroxybutyl}phenoxy)methyl]phosphonate | | Authors: | Lockbaum, G.J, Rusere, L.N, Henes, M, Kosovrasti, K, Lee, S.K, Spielvogel, E, Nalivaika, E.A, Swanstrom, R, KurtYilmaz, N, Schiffer, C.A, Ali, A. | | Deposit date: | 2021-01-14 | | Release date: | 2022-01-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.943 Å) | | Cite: | HIV-1 Protease Inhibitors with a P1 Phosphonate Modification Maintain Potency against Drug Resistant Variants by Increased van der Waals Contacts with Flaps Residues
To Be Published
|
|
7LED
 
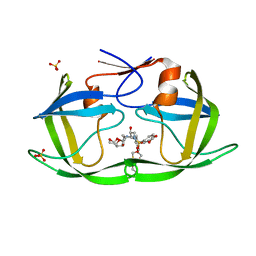 | | HIV-1 Protease WT (NL4-3) in Complex with PU4 (LR2-78) | | Descriptor: | Protease, SULFATE ION, diethyl [(4-{(2S,3R)-4-{[(2H-1,3-benzodioxol-5-yl)sulfonyl][(2S)-2-methylbutyl]amino}-2-[({[(3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl]oxy}carbonyl)amino]-3-hydroxybutyl}phenoxy)methyl]phosphonate | | Authors: | Lockbaum, G.J, Rusere, L.N, Henes, M, Kosovrasti, K, Lee, S.K, Spielvogel, E, Nalivaika, E.A, Swanstrom, R, KurtYilmaz, N, Schiffer, C.A, Ali, A. | | Deposit date: | 2021-01-14 | | Release date: | 2022-01-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.996 Å) | | Cite: | HIV-1 Protease Inhibitors with a P1 Phosphonate Modification Maintain Potency against Drug Resistant Variants by Increased van der Waals Contacts with Flaps Residues
To Be Published
|
|
7LEH
 
 | | HIV-1 Protease WT (NL4-3) in Complex with PU9 (LR2-80) | | Descriptor: | Protease, SULFATE ION, diethyl [(4-{(2S,3R)-4-{[(2H-1,3-benzodioxol-5-yl)sulfonyl](2-ethylbutyl)amino}-2-[({[(3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl]oxy}carbonyl)amino]-3-hydroxybutyl}phenoxy)methyl]phosphonate | | Authors: | Lockbaum, G.J, Rusere, L.N, Henes, M, Kosovrasti, K, Lee, S.K, Spielvogel, E, Nalivaika, E.A, Swanstrom, R, KurtYilmaz, N, Schiffer, C.A, Ali, A. | | Deposit date: | 2021-01-14 | | Release date: | 2022-01-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | HIV-1 Protease Inhibitors with a P1 Phosphonate Modification Maintain Potency against Drug Resistant Variants by Increased van der Waals Contacts with Flaps Residues
To Be Published
|
|
8TJR
 
 | | CRYO-EM STRUCTURE OF HIV-1 BG505DS-SOSIP.664 ENV TRIMER BOUND TO HERH-a.01 FAB | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Antibody HERH-a.01 Heavy Chain, ... | | Authors: | Morano, N.C, Hoyt, F, Hansen, B, Fischer, E, Shapiro, L. | | Deposit date: | 2023-07-24 | | Release date: | 2024-07-31 | | Last modified: | 2025-02-19 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | Potent and broad HIV-1 neutralization in fusion peptide-primed SHIV-infected macaques.
Cell, 187, 2024
|
|
8TJS
 
 | | CRYO-EM STRUCTURE OF HIV-1 BG505DS-SOSIP.664 ENV TRIMER BOUND TO GPZ6-a.01 FAB | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Antibody GPZ6-a.01 Heavy Chain, ... | | Authors: | Morano, N.C, Becker, J.E, Shapiro, L. | | Deposit date: | 2023-07-24 | | Release date: | 2024-07-31 | | Last modified: | 2025-01-15 | | Method: | ELECTRON MICROSCOPY (3.31 Å) | | Cite: | Potent and broad HIV-1 neutralization in fusion peptide-primed SHIV-infected macaques.
Cell, 187, 2024
|
|
8TNU
 
 | | Cryo-EM structure of TRNM-b*01 Fab in complex with HIV-1 Env trimer BG505.DS SOSIP | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BG505 DS-SOSIP Surface protein gp120, ... | | Authors: | Roark, R.S, Morano, N.C, Shapiro, L.S, Kwong, P.D. | | Deposit date: | 2023-08-02 | | Release date: | 2024-08-07 | | Last modified: | 2025-01-15 | | Method: | ELECTRON MICROSCOPY (3.36 Å) | | Cite: | Potent and broad HIV-1 neutralization in fusion peptide-primed SHIV-infected macaques.
Cell, 187, 2024
|
|
