7Q2I
 
 | | Crystal structure of the C-terminal catalytic domain of Plasmodium falciparum CTP:phosphocholine cytidylyltransferase with Tetrahydrofurfurylamine | | Descriptor: | 1-[(2R)-oxolan-2-yl]methanamine, Cholinephosphate cytidylyltransferase, Guanidinium | | Authors: | Duclovel, C, Gelin, M, Krimm, I, Cerdan, R, Guichou, J.-F. | | Deposit date: | 2021-10-25 | | Release date: | 2022-11-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Crystallographic screening using ultra-low-molecular-weight ligands to guide drug design of PfCCT inhibitors
To Be Published
|
|
8EDH
 
 | |
9QWK
 
 | | Crystal structure of S2c-a5b6 TCR in complex with CD1c | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Karuppiah, V, Rangel, V.L. | | Deposit date: | 2025-04-14 | | Release date: | 2025-08-20 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | A CD1c lipid agnostic T cell receptor bispecific engager redirects T cells against CD1c + cells.
Front Immunol, 16, 2025
|
|
9FO3
 
 | | Structure of a gp140 SpyTag-SpyCatcher mi3 nanoparticle including mi3 density only. | | Descriptor: | CAP255 gp140 SpyTag-SpyCatcher mi3,2-dehydro-3-deoxyphosphogluconate aldolase/4-hydroxy-2-oxoglutarate aldolase,2-dehydro-3-deoxyphosphogluconate aldolase/4-hydroxy-2-oxoglutarate aldolase | | Authors: | Woodward, J.D, Malebo, K, Chapman, R. | | Deposit date: | 2024-06-11 | | Release date: | 2024-10-09 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (5.3 Å) | | Cite: | Development of a Two-Component Nanoparticle Vaccine Displaying an HIV-1 Envelope Glycoprotein that Elicits Tier 2 Neutralising Antibodies.
Vaccines (Basel), 12, 2024
|
|
8DR8
 
 | | LRRC8A:C conformation 2 (oblong) top mask | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, Volume-regulated anion channel subunit LRRC8A,Soluble cytochrome b562, Volume-regulated anion channel subunit LRRC8C | | Authors: | Kern, D.M, Brohawn, S.G. | | Deposit date: | 2022-07-20 | | Release date: | 2023-03-08 | | Last modified: | 2023-07-05 | | Method: | ELECTRON MICROSCOPY (3.04 Å) | | Cite: | Structural basis for assembly and lipid-mediated gating of LRRC8A:C volume-regulated anion channels.
Nat.Struct.Mol.Biol., 30, 2023
|
|
9OMV
 
 | | Cryo-EM structure of the C. neoformans lipid flippase Apt1-Cdc50 bound with butyrolactol A in the E2P state | | Descriptor: | (3R,4R,5S)-3,4-dihydroxy-5-[(1R,2R,3S,4S,5R,6R,8E,10E,14E,16Z)-1,2,3,4,5-pentahydroxy-6,20,20-trimethylhenicosa-8,10,14,16-tetraen-1-yl]oxolan-2-one, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Duan, H.D, Li, H. | | Deposit date: | 2025-05-14 | | Release date: | 2025-10-22 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | Butyrolactol A potentiates caspofungin efficacy against resistant fungi via phospholipid flippase inhibition.
Biorxiv, 2025
|
|
6UZP
 
 | |
6VAN
 
 | | Crystal structure of caltubin from the great pond snail | | Descriptor: | 1,2-ETHANEDIOL, Caltubin, EF-hand, ... | | Authors: | Dong, A, Li, A, Zhang, Q, Barszczyk, A, Chern, Y.H, Arrowsmith, C.H, Edwards, A.M, Zhong, Z.P, Tong, Y, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-12-17 | | Release date: | 2020-12-23 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Cell-penetrating caltubin promotes neurite outgrowth and regrowth through calcium-dependent microtubule regulation
to be published
|
|
7Q98
 
 | | MHC Class I A02 Allele presenting NLSALGIFST | | Descriptor: | 1,2-ETHANEDIOL, ASN-LEU-SER-ALA-LEU-GLY-ILE-PHE-SER-THR, Beta-2-microglobulin, ... | | Authors: | Rizkallah, P.J, Sewell, A.K. | | Deposit date: | 2021-11-12 | | Release date: | 2023-02-22 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Targeting of multiple tumor-associated antigens by individual T cell receptors during successful cancer immunotherapy.
Cell, 186, 2023
|
|
7Q9B
 
 | | MHC Class I A02 Allele presenting EAAGIGILTV, in complex with Mel8 TCR | | Descriptor: | 1,2-ETHANEDIOL, Beta-2-microglobulin, GLU-ALA-ALA-GLY-ILE-GLY-ILE-LEU-THR-VAL, ... | | Authors: | Rizkallah, P.J, Sewell, A.K, Wall, A, Fuller, A. | | Deposit date: | 2021-11-12 | | Release date: | 2023-02-22 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.24 Å) | | Cite: | Targeting of multiple tumor-associated antigens by individual T cell receptors during successful cancer immunotherapy.
Cell, 186, 2023
|
|
6VB2
 
 | |
6VB5
 
 | |
6FRC
 
 | |
8SPK
 
 | | Crystal structure of Antarctic PET-degrading enzyme | | Descriptor: | Lipase 1, MALONATE ION | | Authors: | Furtado, A.A, Blazquez-Sanchez, P, Grinen, A, Vargas, J.A, Leonardo, D.A, Sculaccio, S.A, Pereira, H.M, Diez, B, Garratt, R.C, Ramirez-Sarmiento, C.A. | | Deposit date: | 2023-05-03 | | Release date: | 2023-08-23 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Engineering the catalytic activity of an Antarctic PET-degrading enzyme by loop exchange.
Protein Sci., 32, 2023
|
|
5H62
 
 | | Structure of Transferase mutant-C23S,C199S | | Descriptor: | 1,2-ETHANEDIOL, MANGANESE (II) ION, Transferase, ... | | Authors: | Park, J.B, Yoo, Y, Kim, J. | | Deposit date: | 2016-11-10 | | Release date: | 2017-12-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Structural basis for arginine glycosylation of host substrates by bacterial effector proteins.
Nat Commun, 9, 2018
|
|
8EIS
 
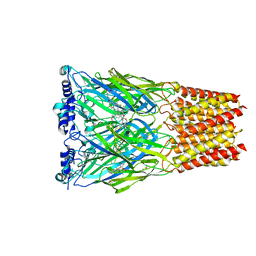 | | Cryo-EM structure of octopus sensory receptor CRT1 | | Descriptor: | 2-[2-[(1~{S},2~{S},4~{S},5'~{R},6~{R},7~{S},8~{R},9~{S},12~{S},13~{R},16~{S})-5',7,9,13-tetramethylspiro[5-oxapentacyclo[10.8.0.0^{2,9}.0^{4,8}.0^{13,18}]icos-18-ene-6,2'-oxane]-16-yl]oxyethyl]propane-1,3-diol, 2-acetamido-2-deoxy-beta-D-glucopyranose, Octopus sensory receptor | | Authors: | Kang, G, Kim, J.J, Allard, C.A.H, Valencia-Montoya, W.A, Bellono, N.W, Hibbs, R.E. | | Deposit date: | 2022-09-15 | | Release date: | 2023-04-12 | | Last modified: | 2025-07-23 | | Method: | ELECTRON MICROSCOPY (2.62 Å) | | Cite: | Structural basis of sensory receptor evolution in octopus.
Nature, 616, 2023
|
|
6MWJ
 
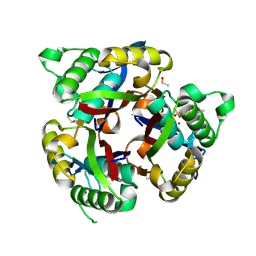 | | Crystal structure of 2C-methyl-D-erythritol 2,4-cyclodiphosphate synthase (IspF) Burkholderia pseudomallei in complex with ligand HGN-0863 | | Descriptor: | 2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase, 5-ACETAMIDO-1,3,4-THIADIAZOLE-2-SULFONAMIDE, DIMETHYL SULFOXIDE, ... | | Authors: | Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2018-10-29 | | Release date: | 2019-11-06 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Analysis of Burkholderia pseudomallei IspF in complex with sulfapyridine, sulfamonomethoxine, ethoxzolamide and acetazolamide
Acta Crystallogr.,Sect.F, 81, 2025
|
|
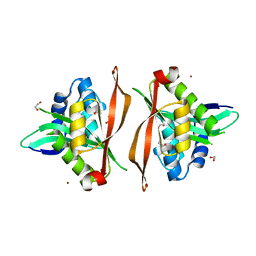
8Q6U
 
 | |
6MM8
 
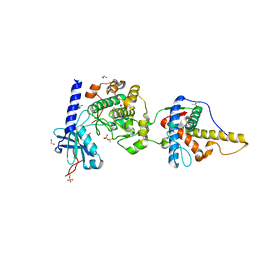 | |
7CSK
 
 | |
7QGJ
 
 | | Apo structure of BIR2 Domain of BIRC2 | | Descriptor: | 1,2-ETHANEDIOL, Baculoviral IAP repeat-containing protein 2, ZINC ION | | Authors: | Kraemer, A, Farges, F, Schwalm, M.P, Saxena, K, Preuss, F, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-12-08 | | Release date: | 2022-02-09 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Apo structure BIR2 Domain of BIRC2
To Be Published
|
|
7QU3
 
 | | X-ray structure of FAD domain of NqrF of Pseudomonas aeruginosa | | Descriptor: | 4-(benzimidazol-1-ylmethyl)benzenecarbonitrile, DIMETHYL SULFOXIDE, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Stegmann, D, Steuber, J, Fritz, G. | | Deposit date: | 2022-01-17 | | Release date: | 2022-02-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Fast fragment- and compound-screening pipeline at the Swiss Light Source.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7QJK
 
 | | Crystal structure of PDE6D in complex with Compound-2 | | Descriptor: | 1,2-ETHANEDIOL, N-(phenylmethyl)pyridin-2-amine, Retinal rod rhodopsin-sensitive cGMP 3',5'-cyclic phosphodiesterase subunit delta, ... | | Authors: | Yelland, T, Ismail, S. | | Deposit date: | 2021-12-16 | | Release date: | 2022-01-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Stabilization of the RAS:PDE6D Complex Is a Novel Strategy to Inhibit RAS Signaling.
J.Med.Chem., 65, 2022
|
|
5HBB
 
 | | Crystal structure of shaft pilin spaA from Lactobacillus rhamnosus GG - E139A mutant | | Descriptor: | 1,2-ETHANEDIOL, Cell surface protein SpaA, SODIUM ION, ... | | Authors: | Chaurasia, P, Pratap, S, von Ossowski, I, Palva, A, Krishnan, V. | | Deposit date: | 2015-12-31 | | Release date: | 2016-07-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | New insights about pilus formation in gut-adapted Lactobacillus rhamnosus GG from the crystal structure of the SpaA backbone-pilin subunit
Sci Rep, 6, 2016
|
|
6FUO
 
 | |
