2LED
 
 | |
2IXA
 
 | | A-zyme, N-acetylgalactosaminidase | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, ALPHA-N-ACETYLGALACTOSAMINIDASE, ... | | Authors: | Sulzenbacher, G, Liu, Q.P, Bourne, Y, Henrissat, B, Clausen, H. | | Deposit date: | 2006-07-07 | | Release date: | 2007-04-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Bacterial Glycosidases for the Production of Universal Red Blood Cells.
Nat.Biotechnol., 25, 2007
|
|
4BNF
 
 | | Crystal structure of S. aureus FabI in complex with NADP and 2- phenoxy-5-propylphenol | | Descriptor: | 2-phenoxy-5-propyl-phenol, ENOYL-[ACYL-CARRIER-PROTEIN] REDUCTASE [NADPH], GLUTAMIC ACID, ... | | Authors: | Schiebel, J, Chang, A, Bommineni, G.R, Tonge, P.J, Kisker, C. | | Deposit date: | 2013-05-15 | | Release date: | 2013-06-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Rational Optimization of Drug-Target Residence Time: Insights from Inhibitor Binding to the S. Aureus Fabi Enzyme-Product Complex.
Biochemistry, 52, 2013
|
|
2IVX
 
 | | Crystal structure of human cyclin T2 at 1.8 A resolution | | Descriptor: | 1,2-ETHANEDIOL, CYCLIN-T2 | | Authors: | Debreczeni, J.E, Bullock, A.N, Fedorov, O, Savitsky, P, Berridge, G, Das, S, Pike, A.C.W, Turnbull, A, Ugochukwu, E, Papagrigoriou, E, Gorrec, F, Sundstrom, M, Edwards, A, Arrowsmith, C, Weigelt, J, von Delft, F, Knapp, S. | | Deposit date: | 2006-06-21 | | Release date: | 2006-07-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The structure of P-TEFb (CDK9/cyclin T1), its complex with flavopiridol and regulation by phosphorylation.
EMBO J., 27, 2008
|
|
4C3F
 
 | |
4CBH
 
 | | Pestivirus NS3 helicase | | Descriptor: | SERINE PROTEASE NS3 | | Authors: | Tortorici, M.A, Duquerroy, S, Kwok, J, Vonrhein, C, Perez, J, Lamp, B, Bricogne, G, Rumenapf, T, Vachette, P, Rey, F.A. | | Deposit date: | 2013-10-14 | | Release date: | 2015-01-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | X-Ray Structure of the Pestivirus Ns3 Helicase and its Conformation in Solution.
J.Virol., 89, 2015
|
|
2LAV
 
 | | NMR solution structure of human Vaccinia-Related Kinase 1 | | Descriptor: | Vaccinia-related kinase 1 | | Authors: | Shin, J, Yoon, H.S. | | Deposit date: | 2011-03-21 | | Release date: | 2011-05-04 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR Solution Structure of Human Vaccinia-related Kinase 1 (VRK1) Reveals the C-terminal Tail Essential for Its Structural Stability and Autocatalytic Activity.
J.Biol.Chem., 286, 2011
|
|
4BZ9
 
 | | Crystal structure of Schistosoma mansoni HDAC8 complexed with J1075 | | Descriptor: | 3-chlorobenzothiophene-2-carbohydroxamic acid, DIMETHYLFORMAMIDE, GLYCEROL, ... | | Authors: | Marek, M, Romier, C. | | Deposit date: | 2013-07-24 | | Release date: | 2013-08-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis for the Inhibition of Histone Deacetylase 8 (Hdac8), a Key Epigenetic Player in the Blood Fluke Schistosoma Mansoni.
Plos Pathog., 9, 2013
|
|
2LEE
 
 | |
4C3C
 
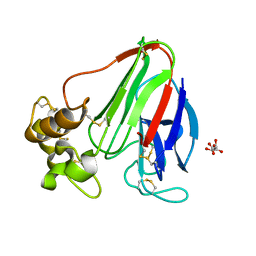 | | Thaumatin refined against hatrx data for time-point 1 | | Descriptor: | L(+)-TARTARIC ACID, THAUMATIN-1 | | Authors: | Yorke, B.A, Beddard, G.S, Owen, R.L, Pearson, A.R. | | Deposit date: | 2013-08-22 | | Release date: | 2014-09-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Time-Resolved Crystallography Using the Hadamard Transform.
Nat.Methods, 11, 2014
|
|
4BW0
 
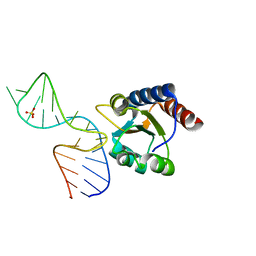 | |
4BZ5
 
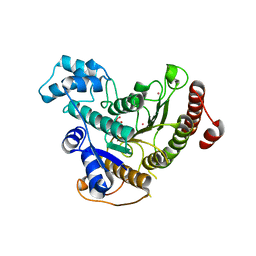 | | Crystal structure of Schistosoma mansoni HDAC8 | | Descriptor: | HISTONE DEACETYLASE 8, L(+)-TARTARIC ACID, POTASSIUM ION, ... | | Authors: | Marek, M, Romier, C. | | Deposit date: | 2013-07-24 | | Release date: | 2013-08-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.785 Å) | | Cite: | Structural Basis for the Inhibition of Histone Deacetylase 8 (Hdac8), a Key Epigenetic Player in the Blood Fluke Schistosoma Mansoni.
Plos Pathog., 9, 2013
|
|
4CND
 
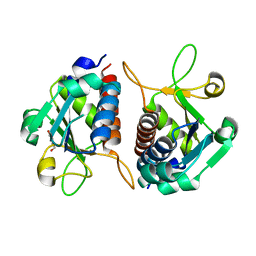 | | Crystal structure of E.coli TrmJ | | Descriptor: | DI(HYDROXYETHYL)ETHER, TRNA (CYTIDINE/URIDINE-2'-O-)-METHYLTRANSFERASE TRMJ | | Authors: | Van Laer, B, Somme, J, Roovers, M, Steyaert, J, Droogmans, L, Versees, W. | | Deposit date: | 2014-01-22 | | Release date: | 2014-07-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Characterization of Two Homologous 2'-O-Methyltransferases Showing Different Specificities for Their tRNA Substrates.
RNA, 20, 2014
|
|
4CNW
 
 | | Surface residue engineering of bovine carbonic anhydrase to an extreme halophilic enzyme for potential application in postcombustion CO2 capture | | Descriptor: | CALCIUM ION, CARBONIC ANHYDRASE 2, ZINC ION | | Authors: | Warden, A, Newman, J, Peat, T.S, Seabrook, S, Williams, M, Dojchinov, G, Haritos, V. | | Deposit date: | 2014-01-25 | | Release date: | 2015-02-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Rational Engineering of a Mesohalophilic Carbonic Anhydrase to an Extreme Halotolerant Biocatalyst.
Nat.Commun., 6, 2015
|
|
4CO5
 
 | |
4CSR
 
 | | High resolution crystal structure of the histone fold dimer (NF-YB)-(NF-YC) | | Descriptor: | GLYCEROL, NUCLEAR TRANSCRIPTION FACTOR Y SUBUNIT BETA, NUCLEAR TRANSCRIPTION FACTOR Y SUBUNIT GAMMA | | Authors: | Gnesutta, N, Cocolo, S, Mantovani, R, Bolognesi, M, Nardini, M. | | Deposit date: | 2014-03-09 | | Release date: | 2015-03-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | High Resolution Crystal Structure of the Histone Fold Dimer (NF-Yb)-(NF-Yc)
To be Published
|
|
4CIH
 
 | | Structure of LntA-K180D-K181D from Listeria monocytogenes | | Descriptor: | LISTERIA NUCLEAR TARGETED PROTEIN A | | Authors: | Lebreton, A, Job, V, Ragon, M, Le Monnier, A, Dessen, A, Cossart, P, Bierne, H. | | Deposit date: | 2013-12-09 | | Release date: | 2014-02-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Structural basis for the inhibition of the chromatin repressor BAHD1 by the bacterial nucleomodulin LntA.
MBio, 5, 2014
|
|
4CNO
 
 | | Structure of the Salmonella typhi Type I dehydroquinase inhibited by a 3-dehydroquinic acid derivative | | Descriptor: | (2R)-2-METHYL-3-DEHYDROQUINIC ACID, 3-DEHYDROQUINATE DEHYDRATASE | | Authors: | Otero, J.M, Llamas-Saiz, A.L, Maneiro, M, Peon, A, Lence, E, Lamb, H, Hawkins, A.R, Gonzalez-Bello, C, van Raaij, M.J. | | Deposit date: | 2014-01-23 | | Release date: | 2014-10-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Insights into substrate binding and catalysis in bacterial type I dehydroquinase.
Biochem. J., 462, 2014
|
|
4CML
 
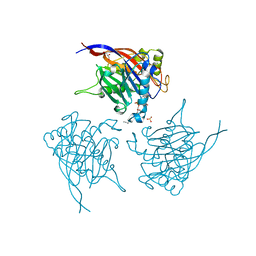 | | Crystal Structure of INPP5B in complex with Phosphatidylinositol 3,4- bisphosphate | | Descriptor: | 1,2-dioctanoyl phosphatidyl epi-inositol (3,4)-bisphosphate, CHLORIDE ION, GLYCEROL, ... | | Authors: | Tresaugues, L, Arrowsmith, C.H, Berglund, H, Bountra, C, Edwards, A.M, Ekblad, T, Flodin, S, Graslund, S, Karlberg, T, Moche, M, Nyman, T, Schuler, H, Silvander, C, Thorsell, A.G, Weigelt, J, Welin, M, Nordlund, P. | | Deposit date: | 2014-01-16 | | Release date: | 2014-04-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis for Phosphoinositide Substrate Recognition, Catalysis, and Membrane Interactions in Human Inositol Polyphosphate 5-Phosphatases.
Structure, 22, 2014
|
|
4CNX
 
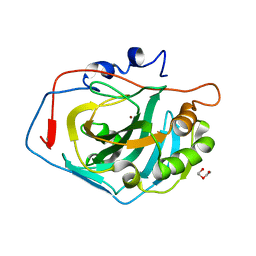 | | Surface residue engineering of bovine carbonic anhydrase to an extreme halophilic enzyme for potential application in postcombustion CO2 capture | | Descriptor: | CARBONIC ANHYDRASE 2, DI(HYDROXYETHYL)ETHER, ZINC ION | | Authors: | Warden, A, Newman, J, Peat, T.S, Seabrook, S, Williams, M, Dojchinov, G, Haritos, V. | | Deposit date: | 2014-01-25 | | Release date: | 2015-02-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.23 Å) | | Cite: | Rational Engineering of a Mesohalophilic Carbonic Anhydrase to an Extreme Halotolerant Biocatalyst.
Nat.Commun., 6, 2015
|
|
2K07
 
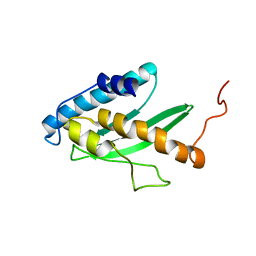 | | Solution NMR structure of human E2-like ubiquitin-fold modifier conjugating enzyme 1 (UFC1). Northeast Structural Genomics Consortium target HR41 | | Descriptor: | Ufm1-conjugating enzyme 1 | | Authors: | Liu, G, Eletsky, A, Atreya, H.S, Aramini, J.M, Xiao, R, Acton, T, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-01-25 | | Release date: | 2008-02-19 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | NMR and X-RAY structures of human E2-like ubiquitin-fold modifier conjugating enzyme 1 (UFC1) reveal structural and functional conservation in the metazoan UFM1-UBA5-UFC1 ubiquination pathway.
J.STRUCT.FUNCT.GENOM., 10, 2009
|
|
4CKX
 
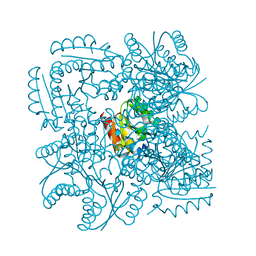 | | Structure of the Mycobacterium tuberculosis Type II Dehydroquinase N12S mutant (Crystal Form 2) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-DEHYDROQUINATE DEHYDRATASE | | Authors: | Otero, J.M, Llamas-Saiz, A.L, Maneiro, M, Peon, A, Sedes, A, Lamb, H, Hawkins, A.R, Gonzalez-Bello, C, van Raaij, M.J. | | Deposit date: | 2014-01-10 | | Release date: | 2015-03-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Investigation of the Dehydratation Mechanism Catalyzed by the Type II Dehydroquinase
To be Published
|
|
2JKU
 
 | | Crystal structure of the N-terminal region of the biotin acceptor domain of human propionyl-CoA carboxylase | | Descriptor: | PROPIONYL-COA CARBOXYLASE ALPHA CHAIN, MITOCHONDRIAL, TETRAETHYLENE GLYCOL | | Authors: | Healy, S, Yue, W.W, Kochan, G, Pilka, E.S, Murray, J.W, Roos, A.K, Filippakopoulos, P, von Delft, F, Arrowsmith, C, Wikstrom, M, Edwards, A, Bountra, C, Gravel, R.A, Oppermann, U. | | Deposit date: | 2008-08-30 | | Release date: | 2008-09-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural impact of human and Escherichia coli biotin carboxyl carrier proteins on biotin attachment.
Biochemistry, 49, 2010
|
|
4CMN
 
 | | Crystal structure of OCRL in complex with a phosphate ion | | Descriptor: | GLYCEROL, INOSITOL POLYPHOSPHATE 5-PHOSPHATASE OCRL-1, MAGNESIUM ION, ... | | Authors: | Tresaugues, L, Moche, M, Arrowsmith, C.H, Berglund, H, Bountra, C, Edwards, A.M, Ekblad, T, Flodin, S, Graslund, S, Karlberg, T, Nyman, T, Schuler, H, Silvander, C, Thorsell, A.G, Weigelt, J, Welin, M, Nordlund, P. | | Deposit date: | 2014-01-16 | | Release date: | 2014-04-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.13 Å) | | Cite: | Structural Basis for Phosphoinositide Substrate Recognition, Catalysis, and Membrane Interactions in Human Inositol Polyphosphate 5-Phosphatases.
Structure, 22, 2014
|
|
4CNV
 
 | | Surface residue engineering of bovine carbonic anhydrase to an extreme halophilic enzyme for potential application in postcombustion CO2 capture | | Descriptor: | CARBONIC ANHYDRASE 2, GLYCEROL, ZINC ION | | Authors: | Warden, A, Newman, J, Peat, T.S, Seabrook, S, Williams, M, Dojchinov, G, Haritos, V. | | Deposit date: | 2014-01-25 | | Release date: | 2015-02-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Rational Engineering of a Mesohalophilic Carbonic Anhydrase to an Extreme Halotolerant Biocatalyst.
Nat.Commun., 6, 2015
|
|
