3V4N
 
 | | The Biochemical and Structural Basis for Inhibition of Enterococcus faecalis HMG-CoA Synthatse, mvaS, by Hymeglusin | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, HMG-CoA synthase | | Authors: | Skaff, D.A, Ramyar, K.X, McWhorter, W.J, Geisbrecht, B.V, Miziorko, H.M. | | Deposit date: | 2011-12-15 | | Release date: | 2012-04-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Biochemical and structural basis for inhibition of Enterococcus faecalis hydroxymethylglutaryl-CoA synthase, mvaS, by hymeglusin.
Biochemistry, 51, 2012
|
|
7S80
 
 | | Crystal structure of iAChSnFR Acetylcholine Sensor precursor binding protein | | Descriptor: | 1,2-ETHANEDIOL, MALONATE ION, iAchSnFR Fluorescent Acetylcholine Sensor precursor binding protein | | Authors: | Fan, C, Borden, P.M, Looger, L.L, Lester, H.A, Rees, D.C. | | Deposit date: | 2021-09-17 | | Release date: | 2021-10-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure, Function, and Application of Bacterial ABC Transporters
Ph.D.Thesis,California Institute of Technology, 2020
|
|
3USN
 
 | | STRUCTURE OF THE CATALYTIC DOMAIN OF HUMAN FIBROBLAST STROMELYSIN-1 INHIBITED WITH THE THIADIAZOLE INHIBITOR IPNU-107859, NMR, 1 STRUCTURE | | Descriptor: | 2-[3-(5-MERCAPTO-[1,3,4]THIADIAZOL-2-YL)-UREIDO]-N-METHYL-3-PHENYL-PROPIONAMIDE, CALCIUM ION, STROMELYSIN-1, ... | | Authors: | Stockman, B.J. | | Deposit date: | 1998-06-18 | | Release date: | 1999-01-20 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structures of stromelysin complexed to thiadiazole inhibitors.
Protein Sci., 7, 1998
|
|
6DVM
 
 | | Crystal structure of Danio rerio histone deacetylase 6 catalytic domain 2 in complex with DDK-122 | | Descriptor: | 1,2-ETHANEDIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 4-(dimethylamino)-N-{[4-(hydroxycarbamoyl)phenyl]methyl}-N-{2-[(4-methylphenyl)amino]-2-oxoethyl}benzamide, ... | | Authors: | Porter, N.J, Christianson, D.W. | | Deposit date: | 2018-06-24 | | Release date: | 2018-08-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Histone Deacetylase 6-Selective Inhibitors and the Influence of Capping Groups on Hydroxamate-Zinc Denticity.
J. Med. Chem., 61, 2018
|
|
3V9O
 
 | |
2P10
 
 | |
8PD1
 
 | | Pseudomonas aeruginosa FabF C164A mutant in complex with N-isopropyl-1H-imidazole-4-carboxamide | | Descriptor: | 3-oxoacyl-[acyl-carrier-protein] synthase 2, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Georgiou, C, Brenk, R, Yadrykhinsky, V, Espeland, L.O. | | Deposit date: | 2023-06-11 | | Release date: | 2024-01-10 | | Last modified: | 2025-05-07 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Towards new antibiotics: P. aeruginosa FabF ligands discovered by crystallographic fragment screening followed by hit expansion.
Eur.J.Med.Chem., 291, 2025
|
|
3MZ2
 
 | |
1M3X
 
 | | Photosynthetic Reaction Center From Rhodobacter Sphaeroides | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN A, ... | | Authors: | Camara-Artigas, A, Brune, D, Allen, J.P. | | Deposit date: | 2002-07-01 | | Release date: | 2002-08-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Interactions between lipids and bacterial reaction centers determined by protein crystallography.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
4NMU
 
 | | Crystal Structure of Thiol-disulfide Oxidoreductase from Bacillus str. 'Ames Ancestor' | | Descriptor: | 1,2-ETHANEDIOL, ACETIC ACID, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Kim, Y, Zhou, M, Shatsman, S, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-11-15 | | Release date: | 2013-12-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Crystal Structure of Thiol-disulfide Oxidoreductase from Bacillus str. 'Ames Ancestor'
To be Published
|
|
6ZR0
 
 | |
8PFZ
 
 | | Pseudomonas aeruginosa FabF C164A mutant in complex with(S)-2-(1H-pyrazole-3-carboxamido)butanoic acid | | Descriptor: | (2~{S})-2-(1~{H}-pyrazol-3-ylcarbonylamino)butanoic acid, 3-oxoacyl-[acyl-carrier-protein] synthase 2, CHLORIDE ION, ... | | Authors: | Georgiou, C, Brenk, R, Yadrykhinsky, V, Espeland, L.O. | | Deposit date: | 2023-06-17 | | Release date: | 2024-01-10 | | Last modified: | 2025-05-07 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Towards new antibiotics: P. aeruginosa FabF ligands discovered by crystallographic fragment screening followed by hit expansion.
Eur.J.Med.Chem., 291, 2025
|
|
6DMS
 
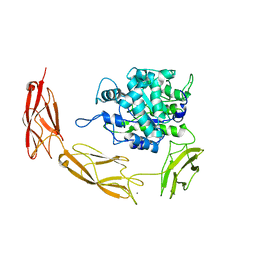 | | Endo-fucoidan hydrolase MfFcnA4_H294Q from glycoside hydrolase family 107 | | Descriptor: | 1,2-ETHANEDIOL, Alpha-1,4-endofucoidanase, CALCIUM ION | | Authors: | Vickers, C, Abe, K, Salama-Alber, O, Boraston, A.B. | | Deposit date: | 2018-06-05 | | Release date: | 2018-10-03 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Endo-fucoidan hydrolases from glycoside hydrolase family 107 (GH107) display structural and mechanistic similarities to alpha-l-fucosidases from GH29.
J. Biol. Chem., 293, 2018
|
|
6ZX9
 
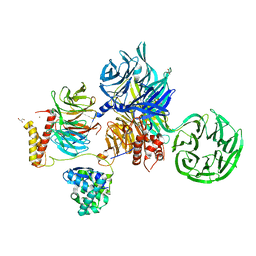 | | Crystal structure of SIV Vpr,fused to T4 lysozyme, isolated from moustached monkey, bound to human DDB1 and human DCAF1 (amino acid residues 1046-1396) | | Descriptor: | DDB1- and CUL4-associated factor 1, DNA damage-binding protein 1, GLYCEROL, ... | | Authors: | Schwefel, D, Banchenko, S. | | Deposit date: | 2020-07-29 | | Release date: | 2021-07-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.519729 Å) | | Cite: | Structural insights into Cullin4-RING ubiquitin ligase remodelling by Vpr from simian immunodeficiency viruses.
Plos Pathog., 17, 2021
|
|
5LEZ
 
 | | Human 20S proteasome complex with Oprozomib in Mg-Acetate at 2.2 Angstrom | | Descriptor: | ACETATE ION, MAGNESIUM ION, PENTAETHYLENE GLYCOL, ... | | Authors: | Schrader, J, Henneberg, F, Mata, R, Tittmann, K, Schneider, T.R, Stark, H, Bourenkov, G, Chari, A. | | Deposit date: | 2016-06-30 | | Release date: | 2016-08-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | The inhibition mechanism of human 20S proteasomes enables next-generation inhibitor design.
Science, 353, 2016
|
|
3VEV
 
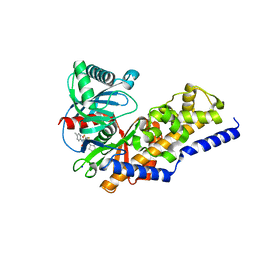 | | Glucokinase in complex with an activator and glucose | | Descriptor: | (2S)-3-cyclopentyl-N-(5-methylpyridin-2-yl)-2-[2-oxo-4-(trifluoromethyl)pyridin-1(2H)-yl]propanamide, Glucokinase, SODIUM ION, ... | | Authors: | Liu, S. | | Deposit date: | 2012-01-09 | | Release date: | 2012-02-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Insights into Mechanism of Glucokinase Activation: OBSERVATION OF MULTIPLE DISTINCT PROTEIN CONFORMATIONS.
J.Biol.Chem., 287, 2012
|
|
2OUA
 
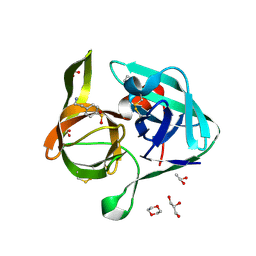 | | Crystal Structure of Nocardiopsis Protease (NAPase) | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, 4-(2-AMINOETHYL)BENZENESULFONYL FLUORIDE, GLYCEROL, ... | | Authors: | Kelch, B.A, Agard, D.A. | | Deposit date: | 2007-02-09 | | Release date: | 2007-02-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural and mechanistic exploration of Acid resistance: kinetic stability facilitates evolution of extremophilic behavior
J.Mol.Biol., 368, 2007
|
|
5FY7
 
 | | Crystal structure of JmjC domain of human histone demethylase UTY in complex with succinate | | Descriptor: | 1,2-ETHANEDIOL, HISTONE DEMETHYLASE UTY, MANGANESE (II) ION, ... | | Authors: | Nowak, R, Krojer, T, Johansson, C, Gileadi, C, Kupinska, K, von Delft, F, Arrowsmith, C.H, Bountra, C, Edwards, A, Oppermann, U. | | Deposit date: | 2016-03-04 | | Release date: | 2016-03-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Crystal Structure of Jmjc Domain of Human Histone Demethylase Uty in Complex with Succinate
To be Published
|
|
6SRU
 
 | | Structure of Ig-like V-type domian of mouse Programmed cell death 1 ligand 1 (PD-L1) | | Descriptor: | Programmed cell death 1 ligand 1 | | Authors: | Magiera-Mularz, K, Sala, D, Grudnik, P, Holak, T.A. | | Deposit date: | 2019-09-06 | | Release date: | 2021-02-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.532 Å) | | Cite: | Human and mouse PD-L1: similar molecular structure, but different druggability profiles.
Iscience, 24, 2021
|
|
3VA8
 
 | | Crystal structure of enolase FG03645.1 (target EFI-502278) from Gibberella zeae PH-1 complexed with magnesium, formate and sulfate | | Descriptor: | FORMIC ACID, MAGNESIUM ION, PROBABLE DEHYDRATASE, ... | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Zencheck, W.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2011-12-29 | | Release date: | 2012-02-01 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of dehydratase FG03645.1 from Gibberella zeae PH-1
To be Published
|
|
6SXX
 
 | | Crystal structure of the bromodomain of human CREBBP in complex with ACA007 | | Descriptor: | (3~{R})-1-ethanoylpyrrolidine-3-carboxylic acid, 1,2-ETHANEDIOL, CREBBP | | Authors: | Xiang, W, Bedi, R.K, Sledz, P, Caflisch, A. | | Deposit date: | 2019-09-26 | | Release date: | 2020-07-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Crystal structure of the bromodomain of human CREBBP in complex with ACA007
To Be Published
|
|
1Z56
 
 | | Co-Crystal Structure of Lif1p-Lig4p | | Descriptor: | DNA ligase IV, Ligase interacting factor 1 | | Authors: | Dore, A.S, Furnham, N, Davies, O.R, Sibanda, B.L, Chirgadze, D.Y, Jackson, S.P, Pellegrini, L, Blundell, T.L. | | Deposit date: | 2005-03-17 | | Release date: | 2006-01-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.92 Å) | | Cite: | Structure of an Xrcc4-DNA ligase IV yeast ortholog complex reveals a novel BRCT interaction mode.
DNA REPAIR, 5, 2006
|
|
3EN5
 
 | | Targeted polypharmacology: crystal structure of the c-Src kinase domain in complex with PP494, a multitargeted kinase inhibitor | | Descriptor: | 1-cyclobutyl-3-(3,4-dimethoxyphenyl)-1H-pyrazolo[3,4-d]pyrimidin-4-amine, Proto-oncogene tyrosine-protein kinase Src | | Authors: | Blair, J.A, Apsel, B, Knight, Z.A, Shokat, K.M. | | Deposit date: | 2008-09-25 | | Release date: | 2008-10-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | Targeted polypharmacology: discovery of dual inhibitors of tyrosine and phosphoinositide kinases.
Nat.Chem.Biol., 4, 2008
|
|
4NUE
 
 | | Crystal structure of the first bromodomain of human BRD4 in complex with MS267 inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 4-[(E)-(2-amino-4-hydroxy-3,5-dimethylphenyl)diazenyl]-N-(pyridin-2-yl)benzenesulfonamide, Bromodomain-containing protein 4 | | Authors: | Plotnikov, A.N, Joshua, J, Zhou, M.-M. | | Deposit date: | 2013-12-03 | | Release date: | 2014-04-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structure-Guided Design of Potent Diazobenzene Inhibitors for the BET Bromodomains
J.Med.Chem., 56, 2013
|
|
3N08
 
 | | Crystal Structure of a Putative PhosphatidylEthanolamine-Binding Protein (PEBP) Homolog CT736 from Chlamydia trachomatis D/UW-3/CX | | Descriptor: | CALCIUM ION, CHLORIDE ION, Putative PhosphatidylEthanolamine-Binding Protein (PEBP) | | Authors: | Brunzelle, J.S, Wawrzak, Z, Onopriyenko, O, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-05-13 | | Release date: | 2010-07-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | 1.25 Angstrom Crystal Structure of a Putative PhosphatidylEthanolamine-Binding Protein (PEBP) Homolog CT736 from Chlamydia trachomatis D/UW-3/CX
To be Published
|
|
