8SPP
 
 | |
5HOD
 
 | | Structure of LHX4 transcription factor complexed with DNA | | Descriptor: | DNA (5'-D(P*AP*CP*CP*TP*AP*AP*TP*TP*AP*GP*GP*CP*GP*TP*AP*AP*TP*TP*AP*G)-3'), DNA (5'-D(P*CP*TP*AP*AP*TP*TP*AP*CP*GP*CP*CP*TP*AP*AP*TP*TP*AP*GP*GP*T)-3'), LIM/homeobox protein Lhx4 | | Authors: | Morgunova, E, Yin, Y, Popov, A, Taipale, J. | | Deposit date: | 2016-01-19 | | Release date: | 2017-02-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.682 Å) | | Cite: | Impact of cytosine methylation on DNA binding specificities of human transcription factors.
Science, 356, 2017
|
|
7BKN
 
 | | Crystal structure of CHK1 complex with adenine | | Descriptor: | ADENINE, SULFATE ION, Serine/threonine-protein kinase Chk1 | | Authors: | Baker, L.M, Surgenor, A.E, Williamson, D.S. | | Deposit date: | 2021-01-16 | | Release date: | 2021-07-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Design and Synthesis of Pyrrolo[2,3- d ]pyrimidine-Derived Leucine-Rich Repeat Kinase 2 (LRRK2) Inhibitors Using a Checkpoint Kinase 1 (CHK1)-Derived Crystallographic Surrogate.
J.Med.Chem., 64, 2021
|
|
6HGN
 
 | |
8SOU
 
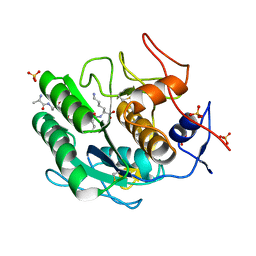 | | Proteinase K Multiconformer Model at 363K | | Descriptor: | ALA-ALA-ALA-SER-VAL-LYS, CALCIUM ION, Proteinase K, ... | | Authors: | Du, S, Wankowicz, S, Yabukarski, F, Doukov, T, Herschlag, D, Fraser, J.S. | | Deposit date: | 2023-04-30 | | Release date: | 2023-08-09 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Refinement of multiconformer ensemble models from multi-temperature X-ray diffraction data.
Methods Enzymol., 688, 2023
|
|
8SQV
 
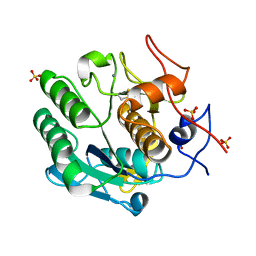 | | Proteinase K Multiconformer Model at 333K | | Descriptor: | CALCIUM ION, Proteinase K, SULFATE ION | | Authors: | Du, S, Wankowicz, S, Yabukarski, F, Doukov, T, Herschlag, D, Fraser, J.S. | | Deposit date: | 2023-05-04 | | Release date: | 2023-08-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | Refinement of multiconformer ensemble models from multi-temperature X-ray diffraction data.
Methods Enzymol., 688, 2023
|
|
8STN
 
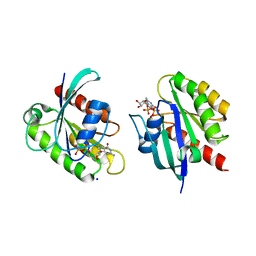 | | Crystal structure of KRAS-G12D/G75A mutant, GDP-bound | | Descriptor: | CHLORIDE ION, GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Tran, T.H, Dharmaiah, S, Simanshu, D.K. | | Deposit date: | 2023-05-10 | | Release date: | 2023-08-23 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Allosteric Regulation of Switch-II Domain Controls KRAS Oncogenicity.
Cancer Res., 83, 2023
|
|
8STM
 
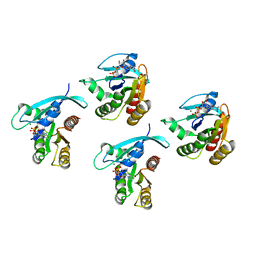 | | Crystal structure of KRAS-G75A mutant, GDP-bound | | Descriptor: | GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION | | Authors: | Tran, T.H, Dharmaiah, S, Simanshu, D.K. | | Deposit date: | 2023-05-10 | | Release date: | 2023-08-23 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Allosteric Regulation of Switch-II Domain Controls KRAS Oncogenicity.
Cancer Res., 83, 2023
|
|
5HPU
 
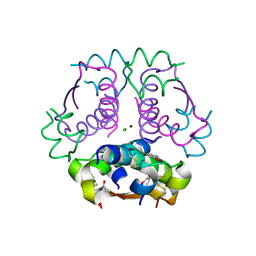 | | Insulin with proline analog HyP at position B28 in the R6 state | | Descriptor: | CHLORIDE ION, Insulin, chain A, ... | | Authors: | Lieblich, S.A, Fang, K.Y, Cahn, J.K.B, Tirrell, D.A. | | Deposit date: | 2016-01-21 | | Release date: | 2017-01-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | 4S-Hydroxylation of Insulin at ProB28 Accelerates Hexamer Dissociation and Delays Fibrillation.
J. Am. Chem. Soc., 139, 2017
|
|
7BHN
 
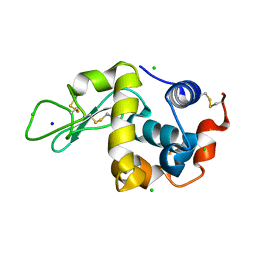 | |
7BHM
 
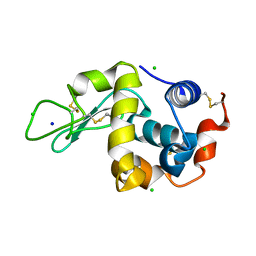 | |
7BHK
 
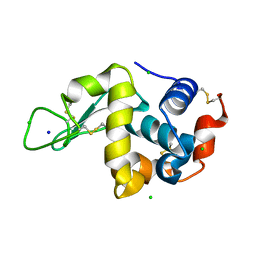 | |
7BHL
 
 | |
5LPV
 
 | |
6GQX
 
 | | KRAS-169 Q61H GPPNHP + CH-2 | | Descriptor: | GTPase KRas, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, ... | | Authors: | Cruz-Migoni, A, Quevedo, C.E, Carr, S.B, Phillips, S.E.V, Rabbitts, T.H. | | Deposit date: | 2018-06-08 | | Release date: | 2019-02-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure-based development of new RAS-effector inhibitors from a combination of active and inactive RAS-binding compounds.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
7BCX
 
 | | The adduct of NAMI-A with Hen Egg White Lysozyme at 8 hours. | | Descriptor: | CHLORIDE ION, DIMETHYL SULFOXIDE, IMIDAZOLE, ... | | Authors: | Chiniadis, L, Giastas, P, Bratsos, I, Papakyriakou, A. | | Deposit date: | 2020-12-21 | | Release date: | 2021-07-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.06 Å) | | Cite: | Insights into the Protein Ruthenation Mechanism by Antimetastatic Metallodrugs: High-Resolution X-ray Structures of the Adduct Formed between Hen Egg-White Lysozyme and NAMI-A at Various Time Points.
Inorg.Chem., 60, 2021
|
|
7BDM
 
 | | The adduct of NAMI-A with Hen Egg White Lysozyme at 98 hours. | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Lysozyme, ... | | Authors: | Chiniadis, L, Giastas, P, Bratsos, I, Papakyriakou, A. | | Deposit date: | 2020-12-22 | | Release date: | 2021-07-28 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.07 Å) | | Cite: | Insights into the Protein Ruthenation Mechanism by Antimetastatic Metallodrugs: High-Resolution X-ray Structures of the Adduct Formed between Hen Egg-White Lysozyme and NAMI-A at Various Time Points.
Inorg.Chem., 60, 2021
|
|
7BD0
 
 | | The adduct of NAMI-A with Hen Egg White Lysozyme at 26 hours. | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, IMIDAZOLE, ... | | Authors: | Chiniadis, L, Giastas, P, Bratsos, I, Papakyriakou, A. | | Deposit date: | 2020-12-21 | | Release date: | 2021-07-28 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.06 Å) | | Cite: | Insights into the Protein Ruthenation Mechanism by Antimetastatic Metallodrugs: High-Resolution X-ray Structures of the Adduct Formed between Hen Egg-White Lysozyme and NAMI-A at Various Time Points.
Inorg.Chem., 60, 2021
|
|
5LHN
 
 | | The catalytic domain of murine urokinase-type plasminogen activator in complex with the allosteric inhibitory nanobody Nb7 | | Descriptor: | 1,2-ETHANEDIOL, Camelid-Derived Antibody Fragment Nb7, SULFATE ION, ... | | Authors: | Kromann-Hansen, T, Lange, E.L, Sorensen, H.P, Ghassabeh, G.H, Huang, M, Jensen, J.K, Muyldermans, S, Declerck, P, Andreasen, P.A. | | Deposit date: | 2016-07-12 | | Release date: | 2017-06-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Discovery of a novel conformational equilibrium in urokinase-type plasminogen activator.
Sci Rep, 7, 2017
|
|
7BCU
 
 | | The adduct of NAMI-A with Hen Egg White Lysozyme at 1.5 hours. | | Descriptor: | CHLORIDE ION, DIMETHYL SULFOXIDE, IMIDAZOLE, ... | | Authors: | Chiniadis, L, Giastas, P, Bratsos, I, Papakyriakou, A. | | Deposit date: | 2020-12-21 | | Release date: | 2021-07-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (0.98 Å) | | Cite: | Insights into the Protein Ruthenation Mechanism by Antimetastatic Metallodrugs: High-Resolution X-ray Structures of the Adduct Formed between Hen Egg-White Lysozyme and NAMI-A at Various Time Points.
Inorg.Chem., 60, 2021
|
|
5HZ5
 
 | |
5H8T
 
 | | Crystal structure of human cellular retinol binding protein 1 in complex with all-trans-retinol | | Descriptor: | RETINOL, Retinol-binding protein 1 | | Authors: | Golczak, M, Arne, J.M, Silvaroli, J.A, Kiser, P.D, Banerjee, S. | | Deposit date: | 2015-12-23 | | Release date: | 2016-03-02 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | Ligand Binding Induces Conformational Changes in Human Cellular Retinol-binding Protein 1 (CRBP1) Revealed by Atomic Resolution Crystal Structures.
J.Biol.Chem., 291, 2016
|
|
5LLZ
 
 | |
8SSP
 
 | | AurA bound to danusertib and activating monobody Mb1 | | Descriptor: | 1,2-ETHANEDIOL, 1,3-PROPANDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ... | | Authors: | Ludewig, H, Kim, C, Kern, D. | | Deposit date: | 2023-05-08 | | Release date: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A biophysical framework for double-drugging kinases.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
5LNN
 
 | | Crystal structure of D1050A mutant of the receiver domain of the histidine kinase CKI1 from Arabidopsis thaliana | | Descriptor: | Histidine kinase CKI1 | | Authors: | Otrusinova, O, Demo, G, Kaderavek, P, Jansen, S, Jasenakova, Z, Pekarova, B, Janda, L, Wimmerova, M, Hejatko, J, Zidek, L. | | Deposit date: | 2016-08-05 | | Release date: | 2017-09-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Conformational dynamics are a key factor in signaling mediated by the receiver domain of a sensor histidine kinase from Arabidopsis thaliana.
J. Biol. Chem., 292, 2017
|
|
