4KYP
 
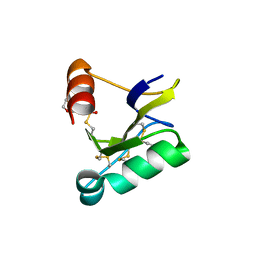 | | Beta-Scorpion Toxin folded in the periplasm of E.coli | | Descriptor: | Beta-insect excitatory toxin Bj-xtrIT, TETRAETHYLENE GLYCOL, TRIETHYLENE GLYCOL | | Authors: | O'Reilly, A.O, Cole, A.R, Lopes, J.L, Lampert, A, Wallace, B.A. | | Deposit date: | 2013-05-29 | | Release date: | 2014-02-12 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Chaperone-mediated native folding of a beta-scorpion toxin in the periplasm of Escherichia coli.
Biochim.Biophys.Acta, 1840, 2014
|
|
4L6T
 
 | | GM1 bound form of the ECX AB5 holotoxin | | Descriptor: | ECXA, ECXB, ZINC ION, ... | | Authors: | Littler, D.R, Ng, N.M, Rossjohn, J, Beddoe, T. | | Deposit date: | 2013-06-12 | | Release date: | 2013-11-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.859 Å) | | Cite: | EcxAB Is a Founding Member of a New Family of Metalloprotease AB5 Toxins with a Hybrid Cholera-like B Subunit.
Structure, 21, 2013
|
|
4L35
 
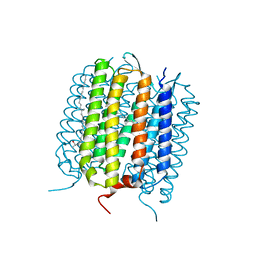 | |
4KV8
 
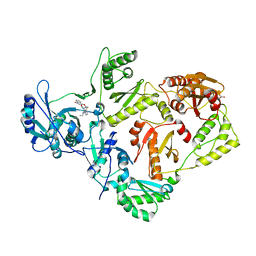 | | Crystal structure of HIV RT in complex with BILR0355BS | | Descriptor: | 11-ethyl-5-methyl-8-[2-(1-oxidanylquinolin-4-yl)oxyethyl]dipyrido[3,2-[1,4]diazepin-6-one, HIV Reverse transcriptase P51, HIV Reverse transcriptase P66, ... | | Authors: | Coulombe, R. | | Deposit date: | 2013-05-22 | | Release date: | 2013-07-31 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | N- versus O-alkylation: Utilizing NMR methods to establish reliable primary structure determinations for drug discovery.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
4KWE
 
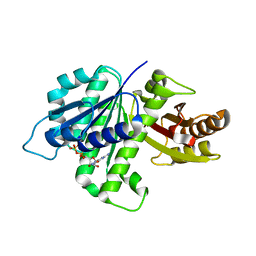 | | GDP-bound, double-stranded, curved FtsZ protofilament structure | | Descriptor: | Cell division protein FtsZ, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Li, Y, Hsin, J, Zhao, L, Cheng, Y, Shang, W, Huang, K.C, Wang, H.W, Ye, S. | | Deposit date: | 2013-05-23 | | Release date: | 2013-07-31 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | FtsZ protofilaments use a hinge-opening mechanism for constrictive force generation
Science, 341, 2013
|
|
4JPX
 
 | |
4JWH
 
 | | Crystal structure of spTrm10(Full length)-SAH complex | | Descriptor: | S-ADENOSYL-L-HOMOCYSTEINE, tRNA (guanine(9)-N1)-methyltransferase | | Authors: | Yan, W, Shao, Z. | | Deposit date: | 2013-03-27 | | Release date: | 2013-10-16 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Crystal structure of tRNA m1G9 methyltransferase Trm10: insight into the catalytic mechanism and recognition of tRNA substrate.
Nucleic Acids Res., 42, 2014
|
|
4JL4
 
 | |
4J9T
 
 | | Crystal structure of a putative, de novo designed unnatural amino acid dependent metalloprotein, northeast structural genomics consortium target OR61 | | Descriptor: | ARSENIC, GLYCEROL, designed unnatural amino acid dependent metalloprotein | | Authors: | Forouhar, F, Lew, S, Seetharaman, J, Mills, J.H, Khare, S.D, Everett, J.K, Baker, D, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2013-02-17 | | Release date: | 2013-03-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Computational design of an unnatural amino Acid dependent metalloprotein with atomic level accuracy.
J.Am.Chem.Soc., 135, 2013
|
|
4JDQ
 
 | |
4JF9
 
 | |
4JJ4
 
 | | Crystal structure of a catalytic mutant of Axe2 (Axe2-D191A), an acetylxylan esterase from Geobacillus stearothermophilus | | Descriptor: | Acetyl xylan esterase, CHLORIDE ION, GLYCEROL | | Authors: | Lansky, S, Alalouf, O, Solomon, V, Alhassid, A, Belrahli, H, Govada, L, Chayan, N.E, Shoham, Y, Shoham, G. | | Deposit date: | 2013-03-07 | | Release date: | 2014-03-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | To be published
To be Published
|
|
4LHF
 
 | | Crystal structure of a DNA binding protein from phage P2 | | Descriptor: | Regulatory protein cox | | Authors: | Berntsson, R.P.-A, Odegrip, R, Sehlen, W, Skaar, K, Svensson, L.M, Massad, T, Haggard-Ljungquist, E, Stenmark, P. | | Deposit date: | 2013-07-01 | | Release date: | 2014-03-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.401 Å) | | Cite: | Structural insight into DNA binding and oligomerization of the multifunctional Cox protein of bacteriophage P2.
Nucleic Acids Res., 42, 2014
|
|
4LHK
 
 | |
4KUM
 
 | | Structure of LSD1-CoREST-Tetrahydrofolate complex | | Descriptor: | (6S)-5,6,7,8-TETRAHYDROFOLATE, CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Luka, Z, Pakhomova, S, Loukachevitch, L.V, Calcutt, M.W, Newcomer, M.E, Wagner, C. | | Deposit date: | 2013-05-22 | | Release date: | 2014-05-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Crystal structure of the histone lysine specific demethylase LSD1 complexed with tetrahydrofolate.
Protein Sci., 23, 2014
|
|
4KK6
 
 | |
4K32
 
 | | Crystal structure of geneticin bound to the leishmanial rRNA A-site | | Descriptor: | GENETICIN, RNA (5'-R(*UP*UP*GP*CP*GP*UP*CP*GP*UP*UP*CP*CP*GP*GP*AP*AP*AP*AP*GP*UP*CP*GP*C)-3') | | Authors: | Shalev, M, Kondo, J, Adir, N, Baasov, T. | | Deposit date: | 2013-04-10 | | Release date: | 2013-07-31 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Identification of the molecular attributes required for aminoglycoside activity against Leishmania.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4KJW
 
 | |
4KPP
 
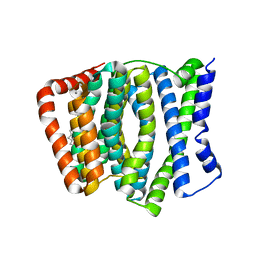 | | Crystal Structure of H+/Ca2+ Exchanger CAX | | Descriptor: | (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, CALCIUM ION, OLEIC ACID, ... | | Authors: | Nishizawa, T, Ishitani, R, Nureki, O. | | Deposit date: | 2013-05-14 | | Release date: | 2013-06-26 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for the counter-transport mechanism of a H+/Ca2+ exchanger.
Science, 341, 2013
|
|
4KKA
 
 | |
4L96
 
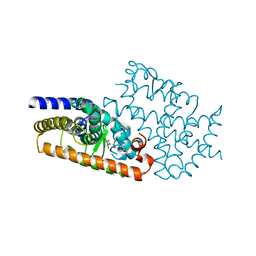 | | Structure of the complex between the F360L PPARgamma mutant and the ligand LT175 (space group I222) | | Descriptor: | (2S)-2-(biphenyl-4-yloxy)-3-phenylpropanoic acid, Peroxisome proliferator-activated receptor gamma | | Authors: | Pochetti, G, Montanari, R, Capelli, D, Chiaraluce, R, Consalvi, V, Pasquo, A, Lori, C, Laghezza, A, Loiodice, F. | | Deposit date: | 2013-06-18 | | Release date: | 2014-06-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Structural basis of the transactivation deficiency of the human PPAR gamma F360L mutant associated with familial partial lipodystrophy.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4L9N
 
 | |
4KY0
 
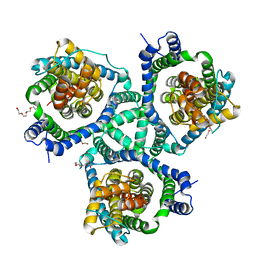 | | Crystal structure of a substrate-free glutamate transporter homologue from Thermococcus kodakarensis | | Descriptor: | Proton/glutamate symporter, SDF family, TETRAETHYLENE GLYCOL | | Authors: | Guskov, A, Jensen, S, Rempel, S, Hanelt, I, Slotboom, D.J. | | Deposit date: | 2013-05-28 | | Release date: | 2013-09-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of a substrate-free aspartate transporter.
Nat.Struct.Mol.Biol., 20, 2013
|
|
4J8Y
 
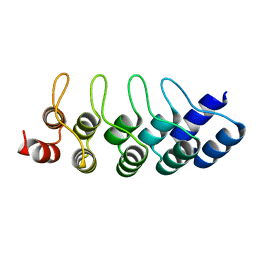 | | E3_5 DARPin D77S mutant | | Descriptor: | DARPin_E3_5_D77S | | Authors: | Seeger, M.A, Gruetter, M.G. | | Deposit date: | 2013-02-15 | | Release date: | 2013-08-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Design, construction, and characterization of a second-generation DARPin library with reduced hydrophobicity.
Protein Sci., 22, 2013
|
|
4JPY
 
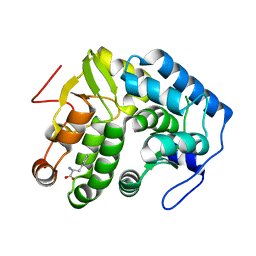 | |
