5WUK
 
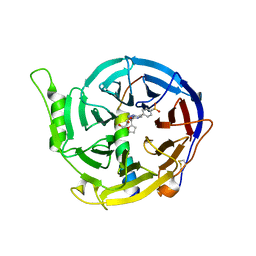 | | Crystal structure of EED [G255D] in complex with EZH2 peptide and EED226 compound | | Descriptor: | GLYCEROL, Histone-lysine N-methyltransferase EZH2, N-(furan-2-ylmethyl)-8-(4-methylsulfonylphenyl)-[1,2,4]triazolo[4,3-c]pyrimidin-5-amine, ... | | Authors: | Chen, Z. | | Deposit date: | 2016-12-19 | | Release date: | 2017-05-03 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Split luciferase-based biosensors for characterizing EED binders
Anal. Biochem., 522, 2017
|
|
6NJZ
 
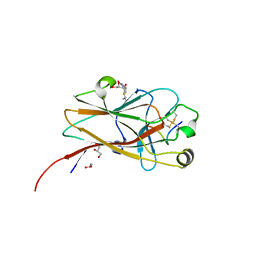 | |
5WMA
 
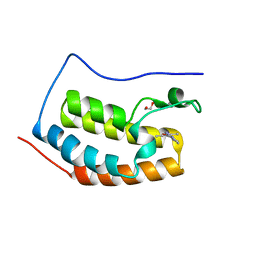 | | N-terminal bromodomain of BRD4 in complex with PLX5981 | | Descriptor: | 1,2-ETHANEDIOL, 5-(3,5-dimethyl-1,2-oxazol-4-yl)-1H-pyrrolo[2,3-b]pyridine, Bromodomain-containing protein 4 | | Authors: | Zhang, Y. | | Deposit date: | 2017-07-28 | | Release date: | 2018-08-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.401 Å) | | Cite: | BRD4 Profiling Identifies Critical Chronic Lymphocytic Leukemia Oncogenic Circuits and Reveals Sensitivity to PLX51107, a Novel Structurally Distinct BET Inhibitor.
Cancer Discov, 8, 2018
|
|
4G17
 
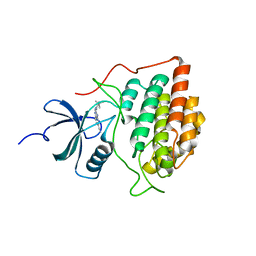 | |
5WZX
 
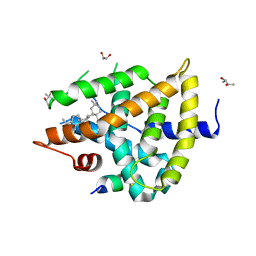 | | Structural basis for a pentacyclic oleanane-type triterpenoid as a ligand of FXR | | Descriptor: | (4aR,6aR,6aS,6bS,8aS,9R,12aR,14bR)-2,2,6a,6b,9,12a-hexamethyl-10-oxidanylidene-1,3,4,5,6,6a,7,8,8a,9,11,12,13,14b-tetradecahydropicene-4a-carboxylic acid, (R,R)-2,3-BUTANEDIOL, Bile acid receptor, ... | | Authors: | Lu, Y, Li, Y. | | Deposit date: | 2017-01-19 | | Release date: | 2018-01-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Identification of an Oleanane-Type Triterpene Hedragonic Acid as a Novel Farnesoid X Receptor Ligand with Liver Protective Effects and Anti-inflammatory Activity
Mol. Pharmacol., 93, 2018
|
|
4G81
 
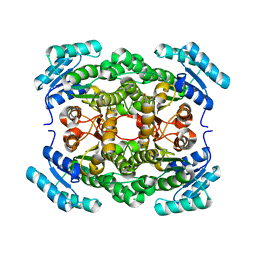 | | Crystal structure of a hexonate dehydrogenase ortholog (target efi-506402 from salmonella enterica, unliganded structure | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Vetting, M.W, Wichelecki, D, Morisco, L.L, Wasserman, S.R, Sojitra, S, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-07-20 | | Release date: | 2012-08-15 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of a hexonate dehydrogenase ortholog (target efi-506402 from salmonella enterica, unliganded structure
To be Published
|
|
1EJN
 
 | | UROKINASE PLASMINOGEN ACTIVATOR B-CHAIN INHIBITOR COMPLEX | | Descriptor: | N-(1-ADAMANTYL)-N'-(4-GUANIDINOBENZYL)UREA, SULFATE ION, UROKINASE-TYPE PLASMINOGEN ACTIVATOR | | Authors: | Sperl, S, Jacob, U, Arroyo de Prada, N, Stuerzebecher, J, Wilhelm, O.G, Bode, W, Magdolen, V, Huber, R, Moroder, L. | | Deposit date: | 2000-04-22 | | Release date: | 2000-05-17 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | (4-aminomethyl)phenylguanidine derivatives as nonpeptidic highly selective inhibitors of human urokinase.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
1EVK
 
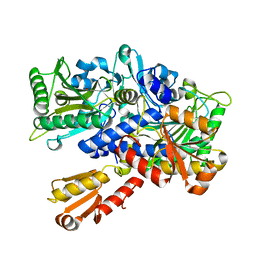 | | CRYSTAL STRUCTURE OF A TRUNCATED FORM OF THREONYL-TRNA SYNTHETASE WITH THE LIGAND THREONINE | | Descriptor: | THREONINE, THREONYL-TRNA SYNTHETASE, ZINC ION | | Authors: | Sankaranarayanan, R, Dock-Bregeon, A.C, Rees, B, Moras, D. | | Deposit date: | 2000-04-20 | | Release date: | 2000-07-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Zinc ion mediated amino acid discrimination by threonyl-tRNA synthetase.
Nat.Struct.Biol., 7, 2000
|
|
5W6K
 
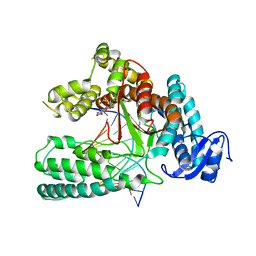 | | Structure of mutant Taq Polymerase incorporating unnatural base pairs Z:P | | Descriptor: | (1R)-1-[6-amino-5-(dihydroxyamino)-2-hydroxypyridin-3-yl]-1,4-anhydro-2-deoxy-5-O-[(S)-hydroxy{[(S)-hydroxy(phosphonooxy)phosphoryl]oxy}phosphoryl]-D-erythro-pentitol, DNA (5'-D(*GP*AP*CP*CP*AP*CP*GP*GP*CP*GP*CP*(DOC))-3'), DNA (5'-D(P*(1WA)P*GP*GP*CP*GP*CP*CP*GP*TP*GP*GP*TP*C)-3'), ... | | Authors: | Singh, I, Georgiadis, M.M. | | Deposit date: | 2017-06-16 | | Release date: | 2018-07-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.339 Å) | | Cite: | Snapshots of an evolved DNA polymerase pre- and post-incorporation of an unnatural nucleotide.
Nucleic Acids Res., 46, 2018
|
|
4FNY
 
 | | Crystal structure of the R1275Q anaplastic lymphoma kinase catalytic domain in complex with a benzoxazole inhibitor | | Descriptor: | ALK tyrosine kinase receptor, N-(4-chlorophenyl)-5-[(6,7-dimethoxyquinolin-4-yl)oxy]-1,3-benzoxazol-2-amine | | Authors: | Whittington, D.A, Epstein, L.F, Chen, H. | | Deposit date: | 2012-06-20 | | Release date: | 2012-08-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | The R1275Q Neuroblastoma Mutant and Certain ATP-competitive Inhibitors Stabilize Alternative Activation Loop Conformations of Anaplastic Lymphoma Kinase.
J.Biol.Chem., 287, 2012
|
|
5LO1
 
 | | HSP90 WITH indazole derivative | | Descriptor: | 1-[2-Amino-4-(1,3-dihydro-isoindole-2-carbonyl)-quinazolin-6-yl]-cyclobutanecarboxylic acid ethylamide, Heat shock protein HSP 90-alpha | | Authors: | Graedler, U, Amaral, M, Schuetz, D. | | Deposit date: | 2016-08-08 | | Release date: | 2017-11-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Ligand Desolvation Steers On-Rate and Impacts Drug Residence Time of Heat Shock Protein 90 (Hsp90) Inhibitors.
J. Med. Chem., 61, 2018
|
|
7A4L
 
 | | PRE-only solution structure of the Iron-Sulfur protein PioC from Rhodopseudomonas palustris TIE-1 | | Descriptor: | IRON/SULFUR CLUSTER, PioC | | Authors: | Trindade, I, Invernici, M, Cantini, F, Louro, R, Piccioli, M. | | Deposit date: | 2020-08-19 | | Release date: | 2020-11-11 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | PRE-driven protein NMR structures: an alternative approach in highly paramagnetic systems.
Febs J., 288, 2021
|
|
7AE6
 
 | | Crystal structure of di-AMPylated HEPN(R102A) toxin | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE MONOPHOSPHATE, HEPN toxin | | Authors: | Tamulaitiene, G, Sasnauskas, G, Songailiene, I, Juozapaitis, J, Siksnys, V. | | Deposit date: | 2020-09-17 | | Release date: | 2020-12-30 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | HEPN-MNT Toxin-Antitoxin System: The HEPN Ribonuclease Is Neutralized by OligoAMPylation.
Mol.Cell, 80, 2020
|
|
1E29
 
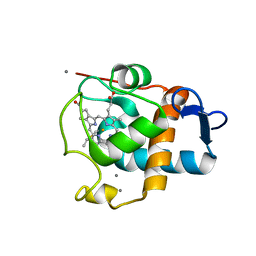 | | PSII associated cytochrome C549 from Synechocystis sp. | | Descriptor: | CALCIUM ION, CYTOCHROME C549, HEME C | | Authors: | Frazao, C, Enguita, F.J, Coelho, R, Sheldrick, G.M. | | Deposit date: | 2000-05-19 | | Release date: | 2001-05-04 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | Crystal Structure of Low-Potential Cytochrome C549 from Synechocystis Sp. Pcc 6803 at 1.21A Resolution
J.Biol.Inorg.Chem., 6, 2001
|
|
5W2S
 
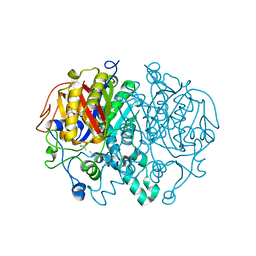 | |
6N8N
 
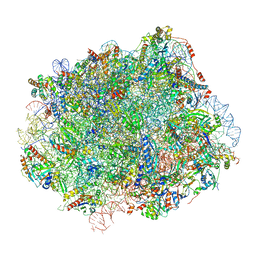 | | Cryo-EM structure of Lsg1-engaged (LE) pre-60S ribosomal subunit | | Descriptor: | 5.8S rRNA, 5S rRNA, 60S ribosomal export protein NMD3, ... | | Authors: | Zhou, Y, Musalgaonkar, S, Johnson, A.W, Taylor, D.W. | | Deposit date: | 2018-11-29 | | Release date: | 2019-03-13 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Tightly-orchestrated rearrangements govern catalytic center assembly of the ribosome.
Nat Commun, 10, 2019
|
|
1DZT
 
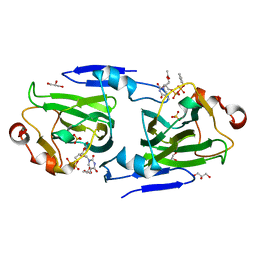 | | RMLC FROM SALMONELLA TYPHIMURIUM | | Descriptor: | 3'-O-ACETYLTHYMIDINE-(5' DIPHOSPHATE PHENYL ESTER), 3'-O-ACETYLTHYMIDINE-5'-DIPHOSPHATE, DTDP-4-DEHYDRORHAMNOSE 3,5-EPIMERASE, ... | | Authors: | Naismith, J.H, Giraud, M.F. | | Deposit date: | 2000-03-07 | | Release date: | 2000-04-18 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Rmlc, the Third Enzyme of Dtdp-L-Rhamnose Pathway, is a New Class of Epimerase.
Nat.Struct.Biol., 7, 2000
|
|
5M0W
 
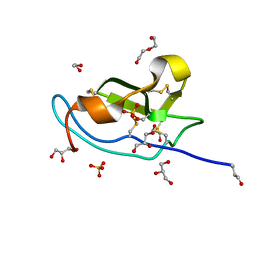 | | N-terminal domain of mouse Shisa 3 | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Lohkamp, B, Ojala, J.R.M. | | Deposit date: | 2016-10-06 | | Release date: | 2017-04-05 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Ab initio solution of macromolecular crystal structures without direct methods.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
1DZ0
 
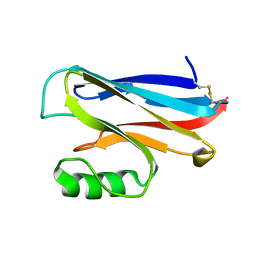 | | REDUCED AZURIN II FROM ALCALIGENES XYLOSOXIDANS | | Descriptor: | AZURIN II, COPPER (I) ION | | Authors: | Dodd, F.E, Abraham, Z.H.L, Eady, R.R, Hasnain, S.S. | | Deposit date: | 2000-02-11 | | Release date: | 2000-07-04 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structures of Oxidised and Reduced Azurin II from Alcaligenes Xylosoxidans at 1.75 Angstoms Resolution
Acta Crystallogr.,Sect.D, 56, 2000
|
|
1E19
 
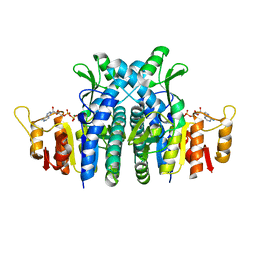 | | Structure of the carbamate kinase-like carbamoyl phosphate synthetase from the hyperthermophilic archaeon Pyrococcus furiosus bound to ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CARBAMATE KINASE, MAGNESIUM ION | | Authors: | Ramon-Maiques, S, Marina, A, Uriarte, M, Fita, I, Rubio, V. | | Deposit date: | 2000-04-28 | | Release date: | 2000-07-04 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The 1.5-A Resolution Crystal Structure of the Carbamate Kinase-Like Carbamoyl Phosphate Synthetase from the Hyperthermophilic Archaeon Pyrococcus Furiosus, Bound to Adp, Confirms that This Thermoestable Enzyme is a Carbamate Kinase, and Provides Insights Into Substrate Binding and Stability in Carbamate Kinases
J.Mol.Biol., 299, 2000
|
|
5M3D
 
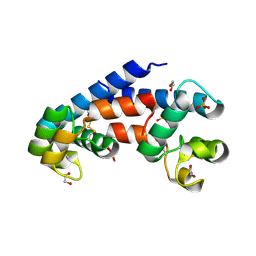 | | Structural tuning of CD81LEL (space group P31) | | Descriptor: | 1,2-ETHANEDIOL, CD81 antigen, PHOSPHATE ION | | Authors: | Cunha, E.S, Sfriso, P, Rojas, A.L, Roversi, P, Hospital, A, Orozco, M, Abrescia, N.G. | | Deposit date: | 2016-10-14 | | Release date: | 2016-12-14 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Mechanism of Structural Tuning of the Hepatitis C Virus Human Cellular Receptor CD81 Large Extracellular Loop.
Structure, 25, 2017
|
|
1DYZ
 
 | | OXIDISED AZURIN II FROM ALCALIGENES XYLOSOXIDANS | | Descriptor: | AZURIN II, COPPER (II) ION | | Authors: | Dodd, F.E, Abraham, Z.H.L, Eady, R.R, Hasnain, S.S. | | Deposit date: | 2000-02-11 | | Release date: | 2000-07-10 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structures of Oxidised and Reduced Azurin II from Alcaligenes Xylosoxidans at 1.75 Angstoms Resolution
Acta Crystallogr.,Sect.D, 56, 2000
|
|
5LV3
 
 | | Crystal structure of mouse CARM1 in complex with ligand LH1561Br | | Descriptor: | 5-[[2-[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]ethylamino]methyl]-4-azanyl-1-[2-(4-bromanylphenoxy)ethyl]pyrimidin-2-one, Histone-arginine methyltransferase CARM1, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Cura, V, Marechal, N, Troffer-Charlier, N, Halby, L, Arimondo, P, Bonnefond, L, Cavarelli, J. | | Deposit date: | 2016-09-12 | | Release date: | 2017-09-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Hijacking DNA methyltransferase transition state analogues to produce chemical scaffolds for PRMT inhibitors.
Philos. Trans. R. Soc. Lond., B, Biol. Sci., 373, 2018
|
|
1DYS
 
 | | Endoglucanase CEL6B from Humicola insolens | | Descriptor: | ENDOGLUCANASE | | Authors: | Davies, G.J, Brzozowski, A.M, Dauter, M, Varrot, A, Schulein, M. | | Deposit date: | 2000-02-08 | | Release date: | 2001-02-08 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure and Function of Humicola Insolens Family 6 Cellulases: Structure of the Endoglucanase, Cel6B, at 1.6 A Resolution
Biochem.J., 348, 2000
|
|
5WEK
 
 | | GluA2 bound to antagonist ZK and GSG1L in digitonin, state 1 | | Descriptor: | Chimera of Glutamate receptor 2,Germ cell-specific gene 1-like protein, {[7-morpholin-4-yl-2,3-dioxo-6-(trifluoromethyl)-3,4-dihydroquinoxalin-1(2H)-yl]methyl}phosphonic acid | | Authors: | Twomey, E.C, Yelshanskaya, M.V, Grassucci, R.A, Frank, J, Sobolevsky, A.I. | | Deposit date: | 2017-07-10 | | Release date: | 2017-08-02 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Channel opening and gating mechanism in AMPA-subtype glutamate receptors.
Nature, 549, 2017
|
|
