6NMP
 
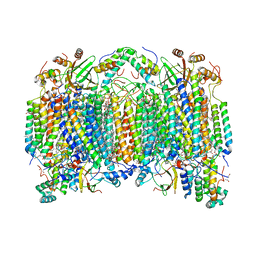 | | SFX structure of oxidized cytochrome c oxidase at room temperature | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, ... | | Authors: | Rousseau, D.L, Yeh, S.-R, Ishigami, I. | | Deposit date: | 2019-01-11 | | Release date: | 2019-03-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Snapshot of an oxygen intermediate in the catalytic reaction of cytochromecoxidase.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
6NNS
 
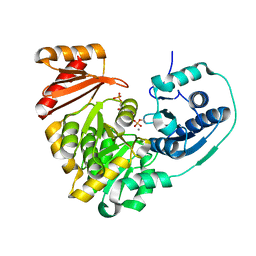 | |
6NPX
 
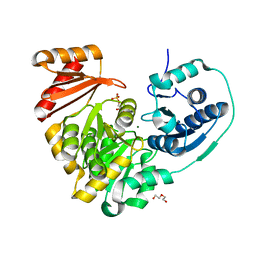 | |
8CJV
 
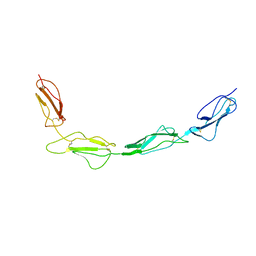 | |
6NNO
 
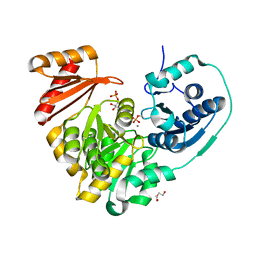 | |
6ZQV
 
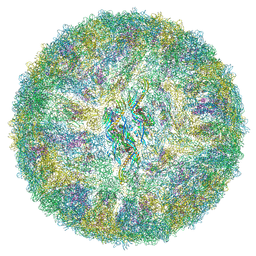 | | Cryo-EM structure of mature Spondweni virus | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Genome polyprotein | | Authors: | Renner, M, Dejnirattisai, W, Carrique, L, Serna Martin, I, Karia, D, Ilca, S.L, Ho, S.F, Kotecha, A, Keown, J.R, Mongkolsapaya, J, Screaton, G.R, Grimes, J.M. | | Deposit date: | 2020-07-10 | | Release date: | 2021-01-20 | | Last modified: | 2021-08-04 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Flavivirus maturation leads to the formation of an occupied lipid pocket in the surface glycoproteins.
Nat Commun, 12, 2021
|
|
8HIP
 
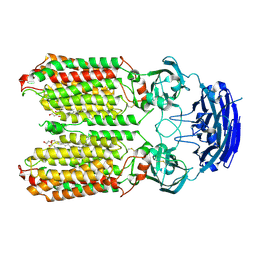 | | dsRNA transporter | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Systemic RNA interference defective protein 1, ZINC ION, ... | | Authors: | Jiang, D.H, Zhang, J.T. | | Deposit date: | 2022-11-21 | | Release date: | 2023-11-29 | | Last modified: | 2024-07-31 | | Method: | ELECTRON MICROSCOPY (2.77 Å) | | Cite: | Structural insights into double-stranded RNA recognition and transport by SID-1.
Nat.Struct.Mol.Biol., 31, 2024
|
|
7A5O
 
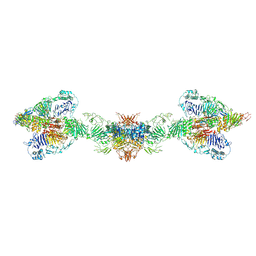 | | Human MUC2 AAs 21-1397 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Javitt, G, Khmelnitsky, L, Albert, L, Elad, N, Ilani, T, Diskin, R, Fass, D. | | Deposit date: | 2020-08-21 | | Release date: | 2020-10-21 | | Last modified: | 2020-11-11 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | Assembly Mechanism of Mucin and von Willebrand Factor Polymers.
Cell, 183, 2020
|
|
8AM9
 
 | | Cryo-EM structure of the proline-rich antimicrobial peptide drosocin bound to the elongating ribosome | | Descriptor: | 16S ribosomal RNA, 2-acetamido-2-deoxy-alpha-D-galactopyranose, 23S ribosomal RNA, ... | | Authors: | Koller, T.O, Morici, M, Wilson, D.N. | | Deposit date: | 2022-08-03 | | Release date: | 2023-03-08 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural basis for translation inhibition by the glycosylated drosocin peptide.
Nat.Chem.Biol., 19, 2023
|
|
8AKN
 
 | | Cryo-EM structure of the proline-rich antimicrobial peptide drosocin bound to the terminating ribosome | | Descriptor: | 16S ribosomal RNA, 2-acetamido-2-deoxy-alpha-D-galactopyranose, 23S ribosomal RNA, ... | | Authors: | Koller, T.O, Morici, M, Wilson, D.N. | | Deposit date: | 2022-07-30 | | Release date: | 2023-03-08 | | Last modified: | 2023-09-06 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | Structural basis for translation inhibition by the glycosylated drosocin peptide.
Nat.Chem.Biol., 19, 2023
|
|
8ANA
 
 | | Cryo-EM structure of the proline-rich antimicrobial peptide drosocin bound to the 50S ribosomal subunit | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-galactopyranose, 23S ribosomal RNA, 50S ribosomal protein L13, ... | | Authors: | Koller, T.O, Morici, M, Wilson, D.N. | | Deposit date: | 2022-08-05 | | Release date: | 2023-03-08 | | Last modified: | 2023-09-06 | | Method: | ELECTRON MICROSCOPY (2.1 Å) | | Cite: | Structural basis for translation inhibition by the glycosylated drosocin peptide.
Nat.Chem.Biol., 19, 2023
|
|
7PGO
 
 | | Crystal Structure of a Class D Carbapenemase_R250A | | Descriptor: | 1-BUTANOL, BROMIDE ION, Beta-lactamase | | Authors: | Zhou, Q, He, Y, Jin, Y. | | Deposit date: | 2021-08-15 | | Release date: | 2022-08-24 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | An Ion-Pair Induced Intermediate Complex Captured in Class D Carbapenemase Reveals Chloride Ion as a Janus Effector Modulating Activity
Acs Cent.Sci., 2023
|
|
7PFN
 
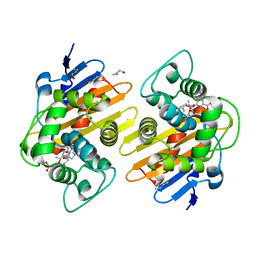 | |
7PEH
 
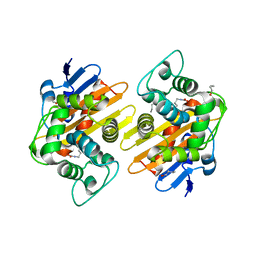 | | Crystal Structure of a Class D Carbapenemase | | Descriptor: | 1-BUTANOL, Beta-lactamase | | Authors: | Zhou, Q, He, Y, Jin, Y. | | Deposit date: | 2021-08-10 | | Release date: | 2022-08-24 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | An Ion-Pair Induced Intermediate Complex Captured in Class D Carbapenemase Reveals Chloride Ion as a Janus Effector Modulating Activity
Acs Cent.Sci., 2023
|
|
7PDA
 
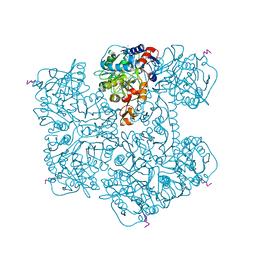 | | Crystal structure of Phenazine 1-carboxylic acid decarboxylase from Mycobacterium fortuitum | | Descriptor: | 1-deoxy-5-O-phosphono-1-(3,3,4,5-tetramethyl-9,11-dioxo-2,3,8,9,10,11-hexahydro-7H-quinolino[1,8-fg]pteridin-12-ium-7-y l)-D-ribitol, MANGANESE (II) ION, SODIUM ION, ... | | Authors: | Gahloth, D, Leys, D. | | Deposit date: | 2021-08-05 | | Release date: | 2022-08-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Crystal structure of Phenazine 1-carboxylic acid decarboxylase from Mycobacterium fortuitum
To Be Published
|
|
7PEP
 
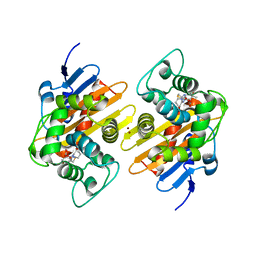 | | Crystal Structure of a Class D Carbapenemase Complexed with Hydrolyzed Imipenem | | Descriptor: | (2R,4S)-2-[(1S,2R)-1-carboxy-2-hydroxypropyl]-4-[(2-{[(Z)-iminomethyl]amino}ethyl)sulfanyl]-3,4-dihydro-2H-pyrrole-5-ca rboxylic acid, 1-BUTANOL, BROMIDE ION, ... | | Authors: | Zhou, Q, He, Y, Jin, Y. | | Deposit date: | 2021-08-11 | | Release date: | 2022-08-24 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | An Ion-Pair Induced Intermediate Complex Captured in Class D Carbapenemase Reveals Chloride Ion as a Janus Effector Modulating Activity
Acs Cent.Sci., 2023
|
|
7PEI
 
 | |
6OUV
 
 | |
7S7K
 
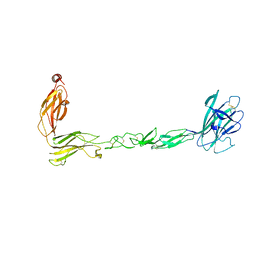 | | Crystal structure of the EphB2 extracellular domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Ephrin type-B receptor 2, ... | | Authors: | Xu, Y, Xu, K, Nikolov, D.B. | | Deposit date: | 2021-09-16 | | Release date: | 2021-10-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | The Ephb2 Receptor Uses Homotypic, Head-to-Tail Interactions within Its Ectodomain as an Autoinhibitory Control Mechanism.
Int J Mol Sci, 22, 2021
|
|
7PSF
 
 | |
8RKE
 
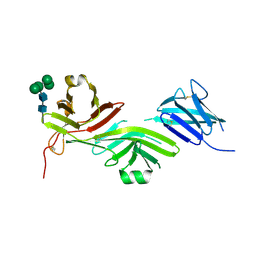 | | Crystal structure of the complete N-terminal region of human ZP2 (hZP2-N1N2N3) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Zona pellucida sperm-binding protein 2, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Fahrenkamp, D, de Sanctis, D, Jovine, L. | | Deposit date: | 2023-12-25 | | Release date: | 2024-03-13 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | ZP2 cleavage blocks polyspermy by modulating the architecture of the egg coat.
Cell, 187, 2024
|
|
8RFG
 
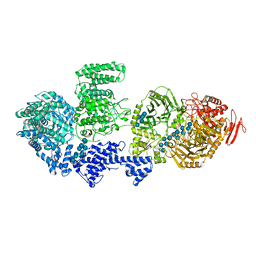 | | CgsiGP3 sample in nanodisc | | Descriptor: | Cyclic beta 1-2 glucan synthetase, beta-D-glucopyranose-(1-2)-beta-D-glucopyranose-(1-2)-beta-D-glucopyranose-(1-2)-beta-D-glucopyranose-(1-2)-beta-D-glucopyranose, beta-D-glucopyranose-(1-2)-beta-D-glucopyranose-(1-2)-beta-D-glucopyranose-(1-2)-beta-D-glucopyranose-(1-2)-beta-D-glucopyranose-(1-2)-beta-D-glucopyranose-(1-2)-beta-D-glucopyranose-(1-2)-beta-D-glucopyranose-(1-2)-beta-D-glucopyranose, ... | | Authors: | Sedzicki, J, Ni, D, Lehmann, F, Stahlberg, H, Dehio, C. | | Deposit date: | 2023-12-12 | | Release date: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.35 Å) | | Cite: | Structure-function analysis of the cyclic beta-1,2-glucan synthase from Agrobacterium tumefaciens.
Nat Commun, 15, 2024
|
|
6ZDG
 
 | | Association of three complexes of largely structurally disordered Spike ectodomain with bound EY6A Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, EY6A heavy chain, EY6A light chain, ... | | Authors: | Duyvesteyn, H.M.E, Zhou, D, Zhao, Y, Fry, E.E, Ren, J, Stuart, D.I. | | Deposit date: | 2020-06-14 | | Release date: | 2020-07-29 | | Last modified: | 2021-12-22 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Structural basis for the neutralization of SARS-CoV-2 by an antibody from a convalescent patient.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6ZCZ
 
 | | Crystal structure of receptor binding domain of SARS-CoV-2 Spike glycoprotein in ternary complex with EY6A Fab and a nanobody. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, EY6A heavy chain, ... | | Authors: | Zhou, D, Zhao, Y, Fry, E.E, Ren, J, Stuart, D.I. | | Deposit date: | 2020-06-12 | | Release date: | 2020-06-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural basis for the neutralization of SARS-CoV-2 by an antibody from a convalescent patient.
Nat.Struct.Mol.Biol., 27, 2020
|
|
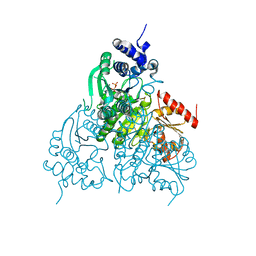
6OUW
 
 | |
