3NCL
 
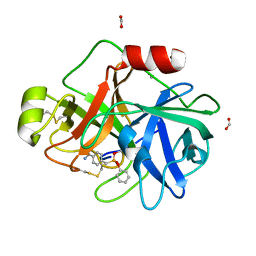 | |
3NI8
 
 | | Crystal Structure of PFC0360w, an HSP90 activator from plasmodium falciparum | | Descriptor: | GLYCEROL, ISOPROPYL ALCOHOL, PFC0360w protein | | Authors: | Wernimont, A.K, Hutchinson, A, Sullivan, H, MacKenzie, F, Kozieradzki, I, Cossar, D, Bochkarev, A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Hui, R, Pizzaro, J.C, Hills, T, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-06-15 | | Release date: | 2010-08-11 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of PFC0360w, an HSP90 activator from plasmodium falciparum
To be Published
|
|
8BVB
 
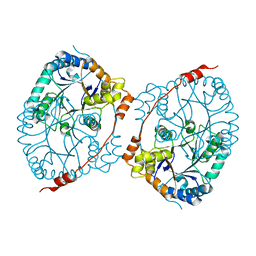 | |
6KSD
 
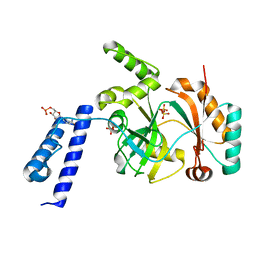 | |
6QYF
 
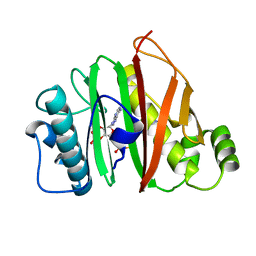 | |
3NUD
 
 | | The structure of 3-deoxy-d-arabino-heptulosonate 7-phosphate synthase from mycobacterium tuberculosis complexed with phenylalanine | | Descriptor: | PHENYLALANINE, PHOSPHATE ION, Probable 3-deoxy-D-arabino-heptulosonate 7-phosphate synthase AroG | | Authors: | Parker, E.J, Jameson, G.B, Jiao, W, Webby, C.J, Baker, E.N, Baker, H.M. | | Deposit date: | 2010-07-06 | | Release date: | 2010-07-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Synergistic allostery, a sophisticated regulatory network for the control of aromatic amino acid biosynthesis in Mycobacterium tuberculosis
J.Biol.Chem., 285, 2010
|
|
6R0E
 
 | |
2YL9
 
 | | INHIBITION OF THE PNEUMOCOCCAL VIRULENCE FACTOR STRH AND MOLECULAR INSIGHTS INTO N-GLYCAN RECOGNITION AND HYDROLYSIS | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BETA-N-ACETYLHEXOSAMINIDASE | | Authors: | Pluvinage, B, Higgins, M.A, Abbott, D.W, Robb, C, Dalia, A.B, Deng, L, Weiser, J.N, Parsons, T.B, Fairbanks, A.J, Vocadlo, D.J, Boraston, A.B. | | Deposit date: | 2011-06-01 | | Release date: | 2011-09-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Inhibition of the Pneumococcal Virulence Factor Strh and Molecular Insights Into N-Glycan Recognition and Hydrolysis.
Structure, 19, 2011
|
|
6KRH
 
 | | Structural basis for domain rotation during adenylation of active site K123 and fragment library screening against NAD+ -dependent DNA ligase from Mycobacterium tuberculosis | | Descriptor: | ADENOSINE MONOPHOSPHATE, BETA-NICOTINAMIDE RIBOSE MONOPHOSPHATE, DNA ligase A, ... | | Authors: | Ramachandran, R, Shukla, A, Afsar, M. | | Deposit date: | 2019-08-21 | | Release date: | 2020-08-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Salt bridges at the subdomain interfaces of the adenylation domain and active-site residues of Mycobacterium tuberculosis NAD + -dependent DNA ligase A (MtbLigA) are important for the initial steps of nick-sealing activity.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
6KVY
 
 | | The structure of EanB/T414A complex with hercynine | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, N,N,N-trimethyl-histidine, ... | | Authors: | Wu, L, Liu, P.H, Zhou, J.H. | | Deposit date: | 2019-09-05 | | Release date: | 2020-09-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | The structure of EanB/T414A complex with hercynine
To Be Published
|
|
6KSC
 
 | |
3NYA
 
 | | Crystal structure of the human beta2 adrenergic receptor in complex with the neutral antagonist alprenolol | | Descriptor: | (2S)-1-[(1-methylethyl)amino]-3-(2-prop-2-en-1-ylphenoxy)propan-2-ol, Beta-2 adrenergic receptor, Lysozyme, ... | | Authors: | Brown, M.A, Wacker, D, Fenalti, G, Katritch, V, Abagyan, R, Cherezov, V, Stevens, R.C, GPCR Network (GPCR) | | Deposit date: | 2010-07-14 | | Release date: | 2010-08-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.16 Å) | | Cite: | Conserved Binding Mode of Human beta(2) Adrenergic Receptor Inverse Agonists and Antagonist Revealed by X-ray Crystallography
J.Am.Chem.Soc., 132, 2010
|
|
3NZN
 
 | | The crystal structure of the Glutaredoxin from Methanosarcina mazei Go1 | | Descriptor: | GLYCEROL, Glutaredoxin, SULFATE ION | | Authors: | Zhang, R, Wu, R, Freeman, L, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-07-16 | | Release date: | 2010-07-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | The crystal structure of the Glutaredoxin from Methanosarcina mazei Go1
To be Published
|
|
8CHH
 
 | |
6KX2
 
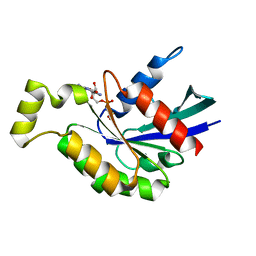 | | Crystal structure of GDP bound RhoA protein | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Transforming protein RhoA | | Authors: | Zhang, H, Luo, C. | | Deposit date: | 2019-09-09 | | Release date: | 2020-08-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.454 Å) | | Cite: | Covalent Inhibitors Allosterically Block the Activation of Rho Family Proteins and Suppress Cancer Cell Invasion.
Adv Sci, 7, 2020
|
|
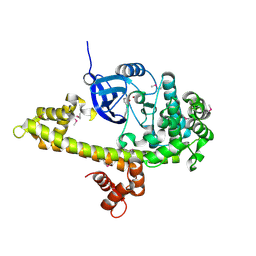
3NYV
 
 | |
6QQC
 
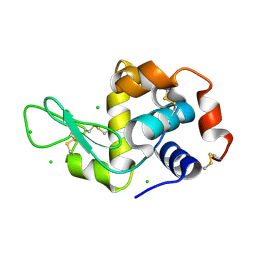 | |
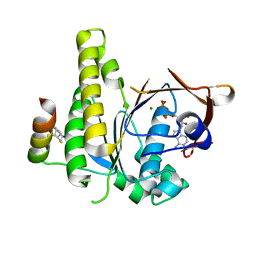
8C3N
 
 | |
8CF4
 
 | |
8CHG
 
 | |
6QPR
 
 | | Rhizomucor miehei lipase propeptide complex, Ser95/Ile96 deletion mutant | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Lipase | | Authors: | Moroz, O.V, Blagova, E, Reiser, V, Saikia, R, Dalal, S, Jorgensen, C.I, Baunsgaard, L, Andersen, B, Svendsen, A, Wilson, K.S. | | Deposit date: | 2019-02-14 | | Release date: | 2019-03-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Novel Inhibitory Function of theRhizomucor mieheiLipase Propeptide and Three-Dimensional Structures of Its Complexes with the Enzyme.
Acs Omega, 4, 2019
|
|
2Y9Y
 
 | | Chromatin Remodeling Factor ISW1a(del_ATPase) | | Descriptor: | IMITATION SWITCH PROTEIN 1 (DEL_ATPASE), ISWI ONE COMPLEX PROTEIN 3 | | Authors: | Yamada, K, Frouws, T.D, Angst, B, Fitzgerald, D.J, DeLuca, C, Schimmele, K, Sargent, D.F, Richmond, T.J. | | Deposit date: | 2011-02-17 | | Release date: | 2011-04-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Structure and Mechanism of the Chromatin Remodelling Factor Isw1A.
Nature, 472, 2011
|
|
2WQJ
 
 | |
6LAA
 
 | | Crystal structure of full-length CYP116B46 from Tepidiphilus thermophilus | | Descriptor: | 1,2-ETHANEDIOL, CARBONATE ION, Cytochrome P450, ... | | Authors: | Zhang, L.L, Ko, T.P, Huang, J.W, Liu, W.D, Chen, C.C, Guo, R.T. | | Deposit date: | 2019-11-12 | | Release date: | 2020-06-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Structural insight into the electron transfer pathway of a self-sufficient P450 monooxygenase.
Nat Commun, 11, 2020
|
|
2WME
 
 | | Crystallographic structure of betaine aldehyde dehydrogenase from Pseudomonas aeruginosa | | Descriptor: | BETA-MERCAPTOETHANOL, BETAINE ALDEHYDE DEHYDROGENASE, GLYCEROL, ... | | Authors: | Gonzalez-Segura, L, Rudino-Pinera, E, Munoz-Clares, R.A, Horjales, E. | | Deposit date: | 2009-06-30 | | Release date: | 2009-08-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Crystal Structure of a Ternary Complex of Betaine Aldehyde Dehydrogenase from Pseudomonas Aeruginosa Provides New Insight Into the Reaction Mechanism and Shows a Novel Binding Mode of the 2'- Phosphate of Nadp(+) and a Novel Cation Binding Site.
J.Mol.Biol., 385, 2009
|
|
