5VYR
 
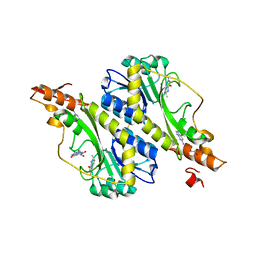 | | Crystal structure of the WbkC formyl transferase from Brucella melitensis | | Descriptor: | (6R)-2-amino-6-methyl-5,6,7,8-tetrahydropteridin-4(3H)-one, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Riegert, A.S, Chantigian, D.P, Thoden, J.B, Holden, H.M. | | Deposit date: | 2017-05-26 | | Release date: | 2017-07-05 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Biochemical Characterization of WbkC, an N-Formyltransferase from Brucella melitensis.
Biochemistry, 56, 2017
|
|
5LTO
 
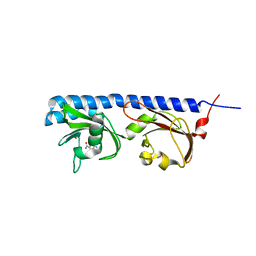 | | Ligand binding domain of Pseudomonas aeruginosa PAO1 amino acid chemoreceptors PctB in complex with L-Gln | | Descriptor: | GLUTAMINE, GLYCEROL, Methyl-accepting chemotaxis protein PctB, ... | | Authors: | Gavira, J.A, Rico-Jimenez, M, Conejero-Muriel, M, Krell, T. | | Deposit date: | 2016-09-07 | | Release date: | 2017-09-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.459 Å) | | Cite: | How Bacterial Chemoreceptors Evolve Novel Ligand Specificities
Mbio, 2020
|
|
5VZ0
 
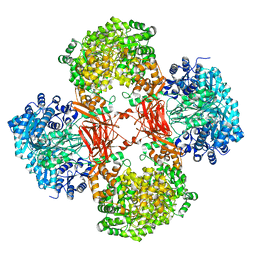 | | Crystal structure of Lactococcus lactis pyruvate carboxylase G746A mutant in complex with cyclic-di-AMP | | Descriptor: | (2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-2,9-bis(6-amino-9H-purin-9-yl)octahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8 ]tetraoxadiphosphacyclododecine-3,5,10,12-tetrol 5,12-dioxide, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Choi, P.H, Tong, L. | | Deposit date: | 2017-05-26 | | Release date: | 2017-08-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and functional studies of pyruvate carboxylase regulation by cyclic di-AMP in lactic acid bacteria.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5W0C
 
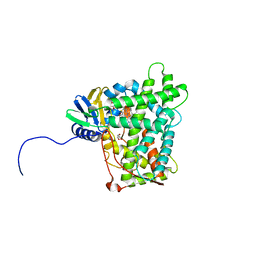 | | Cytochrome P450 (CYP) 2C9 TCA007 Inhibitor Complex | | Descriptor: | Cytochrome P450 2C9, GLYCEROL, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Johnson, E.F, Hsu, M.-H. | | Deposit date: | 2017-05-30 | | Release date: | 2017-09-13 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Determinants of the Inhibition of DprE1 and CYP2C9 by Antitubercular Thiophenes.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
8GS6
 
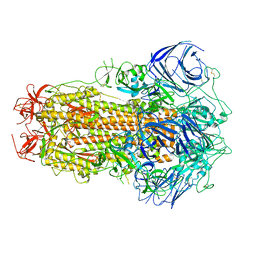 | | Structure of the SARS-CoV-2 BA.2.75 spike glycoprotein (closed state 1) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Anraku, Y, Tabata-Sasaki, K, Kita, S, Fukuhara, H, Maenaka, K, Hashiguchi, T. | | Deposit date: | 2022-09-05 | | Release date: | 2022-10-26 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.86 Å) | | Cite: | Virological characteristics of the SARS-CoV-2 Omicron BA.2.75 variant.
Cell Host Microbe, 30, 2022
|
|
8H11
 
 | | Structure of SARS-CoV-1 Spike Protein with Engineered x1 Disulfide (S370C and D967C), Closed Conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Zhang, X, Li, Z, Liu, Y, Wang, J, Fu, L, Wang, P, He, J, Xiong, X. | | Deposit date: | 2022-09-30 | | Release date: | 2022-11-09 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.72 Å) | | Cite: | Disulfide stabilization reveals conserved dynamic features between SARS-CoV-1 and SARS-CoV-2 spikes.
Life Sci Alliance, 6, 2023
|
|
5W68
 
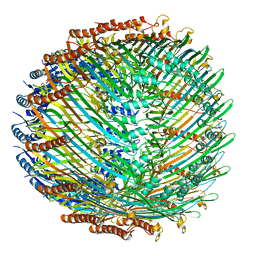 | | Type II secretin from Enteropathogenic Escherichia coli - GspD | | Descriptor: | Putative type II secretion protein | | Authors: | Hay, I.D, Belousoff, M.J, Dunstan, R, Bamert, R, Lithgow, T. | | Deposit date: | 2017-06-16 | | Release date: | 2017-11-15 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure and Membrane Topography of the Vibrio-Type Secretin Complex from the Type 2 Secretion System of Enteropathogenic Escherichia coli.
J. Bacteriol., 200, 2018
|
|
8H0Z
 
 | | Structure of SARS-CoV-1 Spike Protein with Engineered x1 Disulfide (S370C and D967C), Locked-122 Conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BILIVERDINE IX ALPHA, ... | | Authors: | Zhang, X, Li, Z, Liu, Y, Wang, J, Fu, L, Wang, P, He, J, Xiong, X. | | Deposit date: | 2022-09-30 | | Release date: | 2022-11-09 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.99 Å) | | Cite: | Disulfide stabilization reveals conserved dynamic features between SARS-CoV-1 and SARS-CoV-2 spikes.
Life Sci Alliance, 6, 2023
|
|
8H12
 
 | | Structure of SARS-CoV-1 Spike Protein with Engineered x2 Disulfide (G400C and V969C), Locked-2 Conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Zhang, X, Li, Z, Liu, Y, Wang, J, Fu, L, Wang, P, He, J, Xiong, X. | | Deposit date: | 2022-09-30 | | Release date: | 2022-11-09 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.44681 Å) | | Cite: | Disulfide stabilization reveals conserved dynamic features between SARS-CoV-1 and SARS-CoV-2 spikes.
Life Sci Alliance, 6, 2023
|
|
5W55
 
 | |
5M21
 
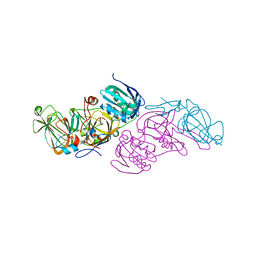 | | Crystal structure of hydroquinone 1,2-dioxygenase from Sphingomonas sp. TTNP3 with 4-hydroxybenzoate bound | | Descriptor: | FE (III) ION, Hydroquinone dioxygenase large subunit, Hydroquinone dioxygenase small subunit, ... | | Authors: | Ferraroni, M, Da Vela, S, Scozzafava, A, Kolvenbach, B, Corvini, P.F.X. | | Deposit date: | 2016-10-11 | | Release date: | 2017-09-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | The crystal structures of native hydroquinone 1,2-dioxygenase from Sphingomonas sp. TTNP3 and of substrate and inhibitor complexes.
Biochim. Biophys. Acta, 1865, 2017
|
|
5W60
 
 | |
5W4D
 
 | | C. japonica N-domain, Selenomethionine mutant | | Descriptor: | 1,2-ETHANEDIOL, 1-METHOXY-2-[2-(2-METHOXY-ETHOXY]-ETHANE, CHLORIDE ION, ... | | Authors: | Aoki, S.T, Bingman, C.A, Kimble, J. | | Deposit date: | 2017-06-10 | | Release date: | 2018-06-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.599 Å) | | Cite: | C. elegans germ granules require both assembly and localized regulators for mRNA repression.
Nat Commun, 12, 2021
|
|
6PQL
 
 | | SBP RafE in complex with raffinose | | Descriptor: | ABC transporter sugar-binding protein, beta-D-fructofuranose-(2-1)-[alpha-D-galactopyranose-(1-6)]alpha-D-glucopyranose | | Authors: | Meier, E.P.W, Boraston, A.B. | | Deposit date: | 2019-07-09 | | Release date: | 2019-10-02 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Molecular analysis of an enigmaticStreptococcus pneumoniaevirulence factor: The raffinose-family oligosaccharide utilization system.
J.Biol.Chem., 294, 2019
|
|
5W5F
 
 | | Cryo-EM structure of the T4 tail tube | | Descriptor: | Tail tube protein gp19 | | Authors: | Zheng, W, Wang, F, Taylor, N.M, Guerrero-Ferreira, R.C, Leiman, P.G, Egelman, E.H. | | Deposit date: | 2017-06-15 | | Release date: | 2017-08-16 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Refined Cryo-EM Structure of the T4 Tail Tube: Exploring the Lowest Dose Limit.
Structure, 25, 2017
|
|
5W61
 
 | |
5W63
 
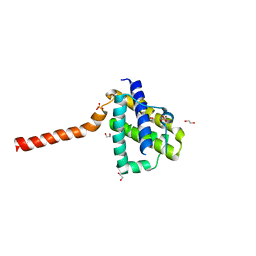 | | Crystal structure of channel catfish BAX | | Descriptor: | 1,2-ETHANEDIOL, Apoptosis regulator bax, SULFATE ION | | Authors: | Robin, A.Y, Colman, P.M, Czabotar, P.E, Luo, C.S. | | Deposit date: | 2017-06-16 | | Release date: | 2018-06-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.436 Å) | | Cite: | Ensemble Properties of Bax Determine Its Function.
Structure, 26, 2018
|
|
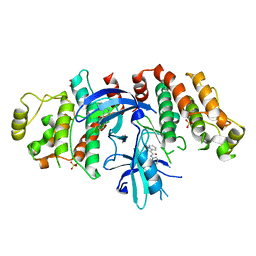
5W5J
 
 | |
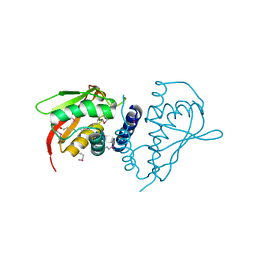
2PWN
 
 | |
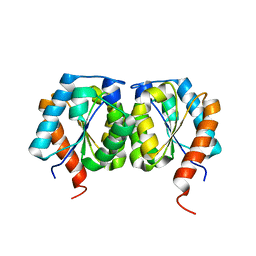
5VTO
 
 | |
5VKG
 
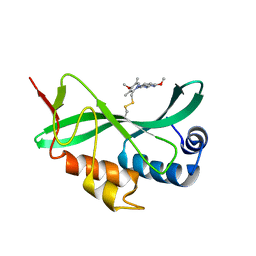 | | Solution-state NMR structural ensemble of human Tsg101 UEV in complex with tenatoprazole | | Descriptor: | 4-methoxy-1-(5-methoxy-3H-imidazo[4,5-b]pyridin-2-yl)-3,5-dimethyl-2-(sulfanylmethyl)pyridin-1-ium, Tumor susceptibility gene 101 protein | | Authors: | Strickland, M, Ehrlich, L.S, Watanabe, S, Khan, M, Strub, M.-P, Luan, C.H, Powell, M.D, Leis, J, Tjandra, N, Carter, C. | | Deposit date: | 2017-04-21 | | Release date: | 2017-11-15 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Tsg101 chaperone function revealed by HIV-1 assembly inhibitors.
Nat Commun, 8, 2017
|
|
5W3N
 
 | | Molecular structure of FUS low sequence complexity domain protein fibrils | | Descriptor: | RNA-binding protein FUS | | Authors: | Murray, D.T, Kato, M, Lin, Y, Thurber, K, Hung, I, McKnight, S, Tycko, R. | | Deposit date: | 2017-06-08 | | Release date: | 2017-09-27 | | Last modified: | 2024-05-15 | | Method: | SOLID-STATE NMR | | Cite: | Structure of FUS Protein Fibrils and Its Relevance to Self-Assembly and Phase Separation of Low-Complexity Domains.
Cell, 171, 2017
|
|
5VF0
 
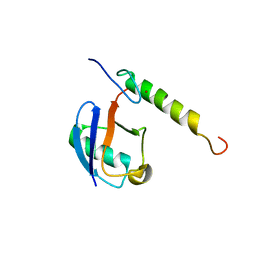 | |
8GZ5
 
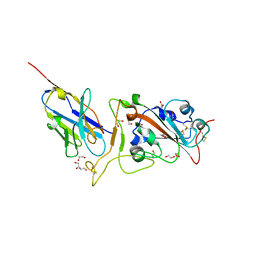 | | Crystal structure of neutralizing VHH P17 in complex with SARS-CoV-2 Alpha variant spike receptor-binding domain | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Nanobody P17, ... | | Authors: | Yamaguchi, K, Anzai, I, Maeda, R, Moriguchi, M, Watanabe, T, Imura, A, Takaori-Kondo, A, Inoue, T. | | Deposit date: | 2022-09-25 | | Release date: | 2022-12-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural insights into the rational design of a nanobody that binds with high affinity to the SARS-CoV-2 spike variant.
J.Biochem., 173, 2023
|
|
5VLN
 
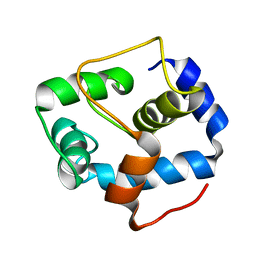 | | NMR structure of the N-domain of troponin C bound to switch region of troponin I | | Descriptor: | Troponin C, slow skeletal and cardiac muscles,Troponin I, cardiac muscle | | Authors: | Cai, F, Hwang, P.M, Sykes, B.D. | | Deposit date: | 2017-04-25 | | Release date: | 2017-05-24 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structures reveal details of small molecule binding to cardiac troponin.
J. Mol. Cell. Cardiol., 101, 2016
|
|
