1GLH
 
 | | CATION BINDING TO A BACILLUS (1,3-1,4)-BETA-GLUCANASE. GEOMETRY, AFFINITY AND EFFECT ON PROTEIN STABILITY | | Descriptor: | 1,3-1,4-BETA-GLUCANASE, SODIUM ION | | Authors: | Keitel, T, Heinemann, U. | | Deposit date: | 1994-11-25 | | Release date: | 1995-02-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Cation binding to a Bacillus (1,3-1,4)-beta-glucanase. Geometry, affinity and effect on protein stability
Eur.J.Biochem., 222, 1994
|
|
3BGZ
 
 | | Human Pim-1 kinase in complex with diphenyl indole inhibitor VX3 | | Descriptor: | 2,3-diphenyl-1H-indole-7-carboxylic acid, Proto-oncogene serine/threonine-protein kinase Pim-1 | | Authors: | Jacobs, M.D. | | Deposit date: | 2007-11-27 | | Release date: | 2007-12-11 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Docking study yields four novel inhibitors of the protooncogene pim-1 kinase.
J.Med.Chem., 51, 2008
|
|
4BG1
 
 | | Three dimensional structure of human gamma-butyrobetaine hydroxylase in complex with 1-(3-Carboxypropyl)-1-methylpyrrolidin-1-ium chloride | | Descriptor: | 1-(3-carboxypropyl)-1-methylpyrrolidin-1-ium, GAMMA-BUTYROBETAINE DIOXYGENASE, HEXANE-1,6-DIAMINE, ... | | Authors: | Tars, K, Leitans, J, Kazaks, A. | | Deposit date: | 2013-03-22 | | Release date: | 2014-03-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Targeting Carnitine Biosynthesis: Discovery of New Inhibitors Against Gamma-Butyrobetaine Hydroxylase.
J.Med.Chem., 57, 2014
|
|
1MAC
 
 | | CRYSTAL STRUCTURE AND SITE-DIRECTED MUTAGENESIS OF BACILLUS MACERANS ENDO-1,3-1,4-BETA-GLUCANASE | | Descriptor: | 1,3-1,4-BETA-D-GLUCAN 4-GLUCANOHYDROLASE, CALCIUM ION | | Authors: | Hahn, M, Heinemann, U. | | Deposit date: | 1994-12-22 | | Release date: | 1995-02-27 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure and site-directed mutagenesis of Bacillus macerans endo-1,3-1,4-beta-glucanase.
J.Biol.Chem., 270, 1995
|
|
2O8Q
 
 | |
4JM9
 
 | | Crystal structure of Cytochrome C Peroxidase W191G-Gateless in complex with 3-amino-1-methylpyridinium | | Descriptor: | 1-METHYL-1,6-DIHYDROPYRIDIN-3-AMINE, Cytochrome c peroxidase, IODIDE ION, ... | | Authors: | Boyce, S.E, Fischer, M, Fish, I. | | Deposit date: | 2013-03-13 | | Release date: | 2013-05-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Blind prediction of charged ligand binding affinities in a model binding site.
J.Mol.Biol., 425, 2013
|
|
2BJK
 
 | | Crystal Analysis of 1-Pyrroline-5-Carboxylate Dehydrogenase from Thermus with bound NAD and citrate. | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, 1-PYRROLINE-5-CARBOXYLATE DEHYDROGENASE, ... | | Authors: | Inagaki, E, Tahirov, T.H. | | Deposit date: | 2005-02-04 | | Release date: | 2006-03-09 | | Last modified: | 2025-10-01 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal Structure of Thermus Thermophilus Delta(1)- Pyrroline-5-Carboxylate Dehydrogenase.
J.Mol.Biol., 362, 2006
|
|
2BHP
 
 | | Crystal Analysis of 1-Pyrroline-5-Carboxylate Dehydrogenase from Thermus with bound NAD. | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, 1-PYRROLINE-5-CARBOXYLATE DEHYDROGENASE, ... | | Authors: | Inagaki, E, Tahirov, T.H. | | Deposit date: | 2005-01-16 | | Release date: | 2006-03-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of Thermus Thermophilus Delta(1)- Pyrroline-5-Carboxylate Dehydrogenase.
J.Mol.Biol., 362, 2006
|
|
7JHI
 
 | | Structure of human beta 1,3-N-acetylglucosaminyltransferase 2 iodide-derivative | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Hao, Y, Huang, X. | | Deposit date: | 2020-07-20 | | Release date: | 2020-11-18 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures and mechanism of human glycosyltransferase beta 1,3-N-acetylglucosaminyltransferase 2 (B3GNT2), an important player in immune homeostasis.
J.Biol.Chem., 296, 2020
|
|
3H98
 
 | | Crystal structure of HCV NS5b 1b with (1,1-dioxo-2H-[1,2,4]benzothiadiazin-3-yl) azolo[1,5-a]pyrimidine derivative | | Descriptor: | GLYCEROL, N-{3-[5-hydroxy-8-(3-methylbutyl)-7-oxo-7,8-dihydroimidazo[1,2-a]pyrimidin-6-yl]-1,1-dioxido-4H-1,2,4-benzothiadiazin-7-yl}methanesulfonamide, RNA-directed RNA polymerase | | Authors: | Wang, G, Lei, H, Wang, X, Das, D, Mackinnon, C, Montalbetti, C.A.G, Mears, R, Gai, X, Bailey, S, Ruhrmund, D, Hooi, L, Misialek, S, Rajagopalan, R, Cheng, R.K.Y, Barker, J.L, Felicetti, B, Stoycheva, A, Buckman, B, Kossen, K, Seiwert, S, Beigelmana, L. | | Deposit date: | 2009-04-30 | | Release date: | 2009-10-13 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | HCV NS5B polymerase inhibitors 2: Synthesis and in vitro activity of (1,1-dioxo-2H-[1,2,4]benzothiadiazin-3-yl) azolo[1,5-a]pyridine and azolo[1,5-a]pyrimidine derivatives.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
7JHN
 
 | | Structure of human beta 1,3-N-acetylglucosaminyltransferase 2 with UDP and trisaccharide GlcNAc-beta1-3Gal-beta1-4GlcNAc | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Hao, Y, Huang, X. | | Deposit date: | 2020-07-21 | | Release date: | 2020-11-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structures and mechanism of human glycosyltransferase beta 1,3-N-acetylglucosaminyltransferase 2 (B3GNT2), an important player in immune homeostasis.
J.Biol.Chem., 296, 2020
|
|
3NOX
 
 | | Crystal structure of human DPP-IV in complex with Sa-(+)-(6-(aminomethyl)-5-(2,4-dichlorophenyl)-7-methylimidazo[1,2-a]pyrimidin-2-yl)(morpholino)methanone | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Dipeptidyl-peptidase 4 (CD26, ... | | Authors: | Klei, H.E. | | Deposit date: | 2010-06-25 | | Release date: | 2010-08-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.338 Å) | | Cite: | Discovery of 6-(Aminomethyl)-5-(2,4-dichlorophenyl)-7-methylimidazo[1,2-a]pyrimidine-2-carboxamides as Potent, Selective Dipeptidyl Peptidase-4 (DPP4) Inhibitors.
J.Med.Chem., 53, 2010
|
|
1K3T
 
 | | Structure of Glycosomal Glyceraldehyde-3-Phosphate Dehydrogenase from Trypanosoma cruzi Complexed with Chalepin, a Coumarin Derivative Inhibitor | | Descriptor: | 6-(1,1-DIMETHYLALLYL)-2-(1-HYDROXY-1-METHYLETHYL)-2,3-DIHYDRO-7H-FURO[3,2-G]CHROMEN-7-ONE, Glyceraldehyde-3-phosphate dehydrogenase | | Authors: | Pavao, F. | | Deposit date: | 2001-10-04 | | Release date: | 2002-06-19 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure of Trypanosoma cruzi glycosomal glyceraldehyde-3-phosphate dehydrogenase complexed with chalepin, a natural product inhibitor, at 1.95 A resolution.
FEBS Lett., 520, 2002
|
|
2CDB
 
 | | Sulfolobus solfataricus Glucose Dehydrogenase 1 in complex with NADP and glucose | | Descriptor: | 1,2-ETHANEDIOL, GLUCOSE 1-DEHYDROGENASE (DHG-1), NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Milburn, C.C, Lamble, H.J, Theodossis, A, Hough, D.W, Danson, M.J, Taylor, G.L. | | Deposit date: | 2006-01-23 | | Release date: | 2006-03-22 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The Structural Basis of Substrate Promiscuity in Glucose Dehydrogenase from the Hyperthermophilic Archaeon Sulfolobus Solfataricus.
J.Biol.Chem., 281, 2006
|
|
2CDC
 
 | | Sulfolobus solfataricus Glucose Dehydrogenase 1 in complex with NADP and Xylose | | Descriptor: | 1,2-ETHANEDIOL, GLUCOSE DEHYDROGENASE GLUCOSE 1-DEHYDROGENASE, DHG-1, ... | | Authors: | Milburn, C.C, Lamble, H.J, Theodossis, A, Hough, D.W, Danson, M.J, Taylor, G.L. | | Deposit date: | 2006-01-23 | | Release date: | 2006-03-22 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The structural basis of substrate promiscuity in glucose dehydrogenase from the hyperthermophilic archaeon Sulfolobus solfataricus.
J. Biol. Chem., 281, 2006
|
|
3CZR
 
 | | Crystal Structure of Human 11-beta-Hydroxysteroid Dehydrogenase (HSD1) in Complex with Arylsulfonylpiperazine Inhibitor | | Descriptor: | (2R)-1-[(4-tert-butylphenyl)sulfonyl]-2-methyl-4-(4-nitrophenyl)piperazine, 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, Corticosteroid 11-beta-dehydrogenase isozyme 1, ... | | Authors: | Wang, Z, Sudom, A, Walker, N.P. | | Deposit date: | 2008-04-29 | | Release date: | 2008-06-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Discovery and Initial SAR of Arylsulfonylpiperazine Inhibitors of 11beta-Hydroxysteroid Dehydrogenase Type 1 (11beta-HSD1)
Bioorg.Med.Chem.Lett., 18, 2008
|
|
2TNF
 
 | | 1.4 A RESOLUTION STRUCTURE OF MOUSE TUMOR NECROSIS FACTOR, TOWARDS MODULATION OF ITS SELECTIVITY AND TRIMERISATION | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ISOPROPYL ALCOHOL, PROTEIN (TUMOR NECROSIS FACTOR ALPHA) | | Authors: | Baeyens, K.J, De Bondt, H.L, Raeymaekers, A, Fiers, W, De Ranter, C.J. | | Deposit date: | 1998-10-12 | | Release date: | 1999-10-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The structure of mouse tumour-necrosis factor at 1.4 A resolution: towards modulation of its selectivity and trimerization.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
1J0D
 
 | | ACC deaminase mutant complexed with ACC | | Descriptor: | 1-aminocyclopropane-1-carboxylate deaminase, N-[3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-Y-LMETHYL]-1-AMINO-CYCLOPROPANECARBOXYLIC ACID | | Authors: | Ose, T, Fujino, A, Yao, M, Honma, M, Tanaka, I. | | Deposit date: | 2002-11-12 | | Release date: | 2003-05-12 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Reaction intermediate structures of 1-aminocyclopropane-1-carboxylate deaminase: insight into PLP-dependent cyclopropane ring-opening reaction
J.BIOL.CHEM., 278, 2003
|
|
3QVT
 
 | | L-myo-inositol 1-phosphate synthase from Archaeoglobus fulgidus wild-type with the intermediate 5-keto 1-phospho glucose | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, GLYCEROL, Myo-inositol-1-phosphate synthase (Ino1), ... | | Authors: | Neelon, K, Roberts, M.F, Stec, B. | | Deposit date: | 2011-02-25 | | Release date: | 2012-01-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a trapped catalytic intermediate suggests that forced atomic proximity drives the catalysis of mIPS.
Biophys.J., 101, 2011
|
|
4IR8
 
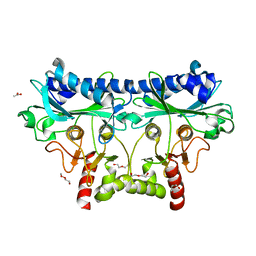 | | 1.85 Angstrom Crystal Structure of Putative Sedoheptulose-1,7 bisphosphatase from Toxoplasma gondii | | Descriptor: | ACETATE ION, DI(HYDROXYETHYL)ETHER, Sedoheptulose-1,7 bisphosphatase, ... | | Authors: | Minasov, G, Ruan, J, Wawrzak, Z, Halavaty, A, Shuvalova, L, Harb, O.S, Ngo, H, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-01-14 | | Release date: | 2013-01-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | 1.85 Angstrom Crystal Structure of Putative Sedoheptulose-1,7 bisphosphatase from Toxoplasma gondii.
TO BE PUBLISHED
|
|
2YHI
 
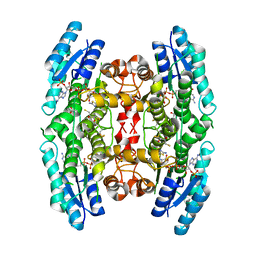 | | Trypanosoma brucei PTR1 in complex with inhibitor WH16 | | Descriptor: | (2R,3S)-1,4-DIMERCAPTOBUTANE-2,3-DIOL, (2S,3S)-1,4-DIMERCAPTOBUTANE-2,3-DIOL, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, ... | | Authors: | Nerini, E, Dawson, A, Hunter, W.N, Costi, M.P. | | Deposit date: | 2011-05-03 | | Release date: | 2012-05-23 | | Last modified: | 2025-10-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Exploiting the 2-Amino-1,3,4-thiadiazole Scaffold To Inhibit Trypanosoma brucei Pteridine Reductase in Support of Early-Stage Drug Discovery.
ACS Omega, 2, 2017
|
|
1RF9
 
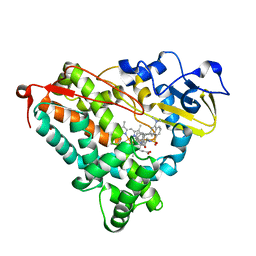 | | Crystal structure of cytochrome P450-cam with a fluorescent probe D-4-AD (Adamantane-1-carboxylic acid-5-dimethylamino-naphthalene-1-sulfonylamino-butyl-amide) | | Descriptor: | 1,2-ETHANEDIOL, ADAMANTANE-1-CARBOXYLIC ACID-5-DIMETHYLAMINO-NAPHTHALENE-1-SULFONYLAMINO-BUTYL-AMIDE, Cytochrome P450-cam, ... | | Authors: | Hays, A.-M.A, Dunn, A.R, Gray, H.B, Stout, C.D, Goodin, D.B. | | Deposit date: | 2003-11-07 | | Release date: | 2004-11-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Conformational States of Cytochrome P450cam Revealed by Trapping of synthetic Molecular Wires
J.Mol.Biol., 344, 2004
|
|
3QQP
 
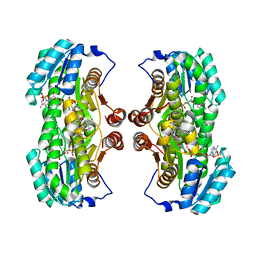 | | Crystal Structure of 11beta-Hydroxysteroid Dehydrogenase 1 (11b-HSD1) in Complex with Urea Inhibitor | | Descriptor: | 3,4-dihydroquinolin-1(2H)-yl[4-(1H-imidazol-5-yl)piperidin-1-yl]methanone, Corticosteroid 11-beta-dehydrogenase isozyme 1, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Loenze, P, Schimanski-Breves, S, Engel, C.K. | | Deposit date: | 2011-02-16 | | Release date: | 2012-02-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Pyrrolidine-pyrazole ureas as potent and selective inhibitors of 11beta-hydroxysteroid-dehydrogenase type 1.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
2YHU
 
 | | Trypanosoma brucei PTR1 in complex with inhibitor WHF30 | | Descriptor: | 3-(5-AMINO-1,3,4-THIADIAZOL-2-YL)-1-THIOPHEN-2-YLPROPAN-1-ONE, ACETATE ION, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Nerini, E, Dawson, A, Hunter, W.N, Costi, M.P. | | Deposit date: | 2011-05-06 | | Release date: | 2012-05-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Exploiting the 2-Amino-1,3,4-thiadiazole Scaffold To Inhibit Trypanosoma brucei Pteridine Reductase in Support of Early-Stage Drug Discovery.
ACS Omega, 2, 2017
|
|
3B9F
 
 | | 1.6 A structure of the PCI-thrombin-heparin complex | | Descriptor: | 2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose, GLYCEROL, Plasma serine protease inhibitor, ... | | Authors: | Li, W, Adams, T.E, Huntington, J.A. | | Deposit date: | 2007-11-05 | | Release date: | 2008-04-22 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Molecular basis of thrombin recognition by protein C inhibitor revealed by the 1.6-A structure of the heparin-bridged complex.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
