3K2W
 
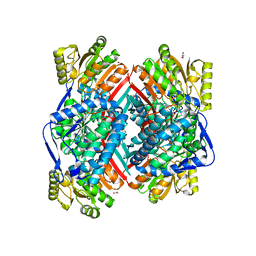 | | CRYSTAL STRUCTURE OF betaine-aldehyde dehydrogenase FROM Pseudoalteromonas atlantica T6c | | Descriptor: | ACETATE ION, Betaine-aldehyde dehydrogenase, CHLORIDE ION, ... | | Authors: | Patskovsky, Y, Toro, R, Rutter, M, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-09-30 | | Release date: | 2009-10-20 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | CRYSTAL STRUCTURE OF betaine-aldehyde dehydrogenase FROM Pseudoalteromonas atlantica
To be Published
|
|
3J9Z
 
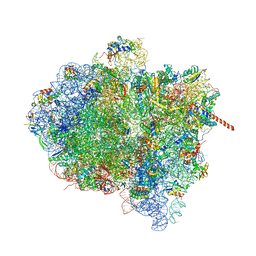 | | Activation of GTP Hydrolysis in mRNA-tRNA Translocation by Elongation Factor G | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Li, W, Liu, Z, Koripella, R.K, Langlois, R, Sanyal, S, Frank, J. | | Deposit date: | 2015-03-27 | | Release date: | 2015-07-01 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Activation of GTP hydrolysis in mRNA-tRNA translocation by elongation factor G.
Sci Adv, 1, 2015
|
|
3JAI
 
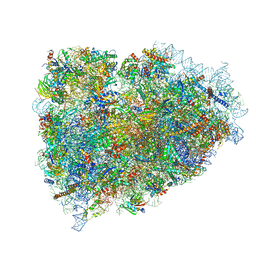 | | Structure of a mammalian ribosomal termination complex with ABCE1, eRF1(AAQ), and the UGA stop codon | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA, 5.8S ribosomal RNA, ... | | Authors: | Brown, A, Shao, S, Murray, J, Hegde, R.S, Ramakrishnan, V. | | Deposit date: | 2015-06-10 | | Release date: | 2015-08-12 | | Last modified: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (3.65 Å) | | Cite: | Structural basis for stop codon recognition in eukaryotes.
Nature, 524, 2015
|
|
3JAJ
 
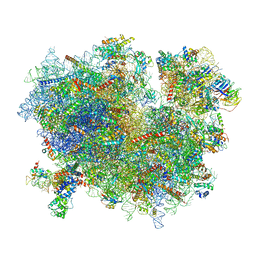 | |
3K2B
 
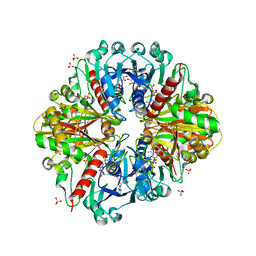 | | Crystal structure of photosynthetic A4 isoform glyceraldehyde-3-phosphate dehydrogenase complexed with NAD, from Arabidopsis thaliana | | Descriptor: | Glyceraldehyde-3-phosphate dehydrogenase A, chloroplastic, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Fermani, S, Falini, G, Thumiger, A, Sparla, F, Marri, L, Trost, P. | | Deposit date: | 2009-09-29 | | Release date: | 2010-06-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of photosynthetic glyceraldehyde-3-phosphate dehydrogenase (isoform A4) from Arabidopsis thaliana in complex with NAD
Acta Crystallogr.,Sect.F, 66, 2010
|
|
3J77
 
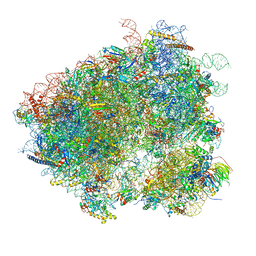 | | Structures of yeast 80S ribosome-tRNA complexes in the rotated and non-rotated conformations (Class II - rotated ribosome with 1 tRNA) | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 40S ribosomal protein S0, ... | | Authors: | Svidritskiy, E, Brilot, A.F, Koh, C.S, Grigorieff, N, Korostelev, A.A. | | Deposit date: | 2014-05-29 | | Release date: | 2014-08-06 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (6.2 Å) | | Cite: | Structures of Yeast 80S Ribosome-tRNA Complexes in the Rotated and Nonrotated Conformations.
Structure, 22, 2014
|
|
3JAH
 
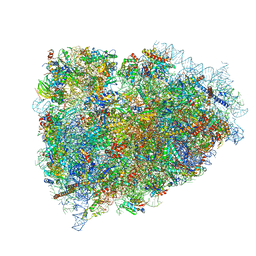 | | Structure of a mammalian ribosomal termination complex with ABCE1, eRF1(AAQ), and the UAG stop codon | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA, 5.8S ribosomal RNA, ... | | Authors: | Brown, A, Shao, S, Murray, J, Hegde, R.S, Ramakrishnan, V. | | Deposit date: | 2015-06-10 | | Release date: | 2015-08-12 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.45 Å) | | Cite: | Structural basis for stop codon recognition in eukaryotes.
Nature, 524, 2015
|
|
3JC1
 
 | | Electron cryo-microscopy of the IST1-CHMP1B ESCRT-III copolymer | | Descriptor: | Charged multivesicular body protein 1b, Increased Sodium Tolerance 1 (IST1) | | Authors: | McCullough, J, Clippinger, A.K, Talledge, N, Skowyra, M.L, Saunders, M.G, Naismith, T.V, Colf, L.A, Afonine, P, Arthur, C, Sundquist, W.I, Hanson, P.I, Frost, A. | | Deposit date: | 2015-11-09 | | Release date: | 2015-12-16 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structure and membrane remodeling activity of ESCRT-III helical polymers.
Science, 350, 2015
|
|
3JCO
 
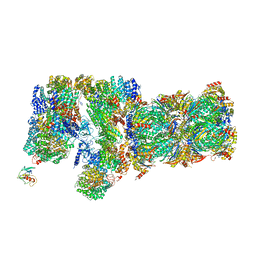 | | Structure of yeast 26S proteasome in M1 state derived from Titan dataset | | Descriptor: | 26S protease regulatory subunit 4 homolog, 26S protease regulatory subunit 6A, 26S protease regulatory subunit 6B homolog, ... | | Authors: | Luan, B, Huang, X.L, Wu, J.P, Shi, Y.G, Wang, F. | | Deposit date: | 2016-01-06 | | Release date: | 2016-06-15 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Structure of an endogenous yeast 26S proteasome reveals two major conformational states.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
3JCT
 
 | | Cryo-em structure of eukaryotic pre-60S ribosomal subunits | | Descriptor: | 60S ribosomal protein L11-A, 60S ribosomal protein L13-A, 60S ribosomal protein L14-A, ... | | Authors: | Wu, S, Kumcuoglu, B, Yan, K.G, Brown, H, Zhang, Y.X, Tan, D, Gamalinda, M, Yuan, Y, Li, Z.F, Jakovljevic, J, Ma, C.Y, Lei, J.L, Dong, M.Q, Woolford Jr, J.L, Gao, N. | | Deposit date: | 2016-03-09 | | Release date: | 2016-06-01 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.08 Å) | | Cite: | Diverse roles of assembly factors revealed by structures of late nuclear pre-60S ribosomes
Nature, 534, 2016
|
|
3K09
 
 | | Crystal structure of the phosphorylation-site mutant S431D of the KaiC circadian clock protein | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Circadian clock protein kinase kaiC, MAGNESIUM ION | | Authors: | Pattanayek, R, Egli, M, Pattanayek, S. | | Deposit date: | 2009-09-24 | | Release date: | 2010-03-31 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structures of KaiC Circadian Clock Mutant Proteins: A New Phosphorylation Site at T426 and Mechanisms of Kinase, ATPase and Phosphatase.
Plos One, 4, 2009
|
|
3K4M
 
 | | Pyranose 2-oxidase Y456W mutant in complex with 2FG | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-deoxy-2-fluoro-beta-D-glucopyranose, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Divne, C, Tan, T.C. | | Deposit date: | 2009-10-05 | | Release date: | 2010-05-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Importance of the gating segment in the substrate-recognition loop of pyranose 2-oxidase.
Febs J., 277, 2010
|
|
3JW7
 
 | | Crystal structure of Dipeptide Epimerase from Enterococcus faecalis V583 complexed with Mg and dipeptide L-Ile-L-Tyr | | Descriptor: | Dipeptide Epimerase, GLYCEROL, ISOLEUCINE, ... | | Authors: | Fedorov, A.A, Fedorov, E.V, Imker, H.J, Sakai, A, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2009-09-18 | | Release date: | 2010-08-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Homology models guide discovery of diverse enzyme specificities among dipeptide epimerases in the enolase superfamily.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3JAM
 
 | | CryoEM structure of 40S-eIF1A-eIF1 complex from yeast | | Descriptor: | 18S rRNA, MAGNESIUM ION, RACK1, ... | | Authors: | Llacer, J.L, Hussain, T, Ramakrishnan, V. | | Deposit date: | 2015-06-17 | | Release date: | 2015-08-12 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.46 Å) | | Cite: | Conformational Differences between Open and Closed States of the Eukaryotic Translation Initiation Complex.
Mol.Cell, 59, 2015
|
|
3K0D
 
 | | Crystal Structure of CNG mimicking NaK mutant, NaK-ETPP, K+ complex | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, POTASSIUM ION, Potassium channel protein NaK | | Authors: | Jiang, Y, Derebe, M.G. | | Deposit date: | 2009-09-24 | | Release date: | 2011-01-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural studies of ion permeation and Ca2+ blockage of a bacterial channel mimicking the cyclic nucleotide-gated channel pore.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3JB9
 
 | | Cryo-EM structure of the yeast spliceosome at 3.6 angstrom resolution | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Yan, C, Hang, J, Wan, R, Huang, M, Wong, C, Shi, Y. | | Deposit date: | 2015-08-09 | | Release date: | 2015-09-23 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structure of a yeast spliceosome at 3.6-angstrom resolution
Science, 349, 2015
|
|
3JCJ
 
 | | Structures of ribosome-bound initiation factor 2 reveal the mechanism of subunit association | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Sprink, T, Ramrath, D.J.F, Yamamoto, H, Yamamoto, K, Loerke, J, Ismer, J, Hildebrand, P.W, Scheerer, P, Buerger, J, Mielke, T, Spahn, C.M.T. | | Deposit date: | 2015-12-18 | | Release date: | 2016-03-09 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structures of ribosome-bound initiation factor 2 reveal the mechanism of subunit association.
Sci Adv, 2, 2016
|
|
1BHQ
 
 | | MAC-1 I DOMAIN CADMIUM COMPLEX | | Descriptor: | ACETYL GROUP, CADMIUM ION, CD11B | | Authors: | Baldwin, E.T. | | Deposit date: | 1998-06-10 | | Release date: | 1998-11-04 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Cation binding to the integrin CD11b I domain and activation model assessment
Structure, 6, 1998
|
|
1BHO
 
 | | MAC-1 I DOMAIN MAGNESIUM COMPLEX | | Descriptor: | CD11B, MAGNESIUM ION | | Authors: | Baldwin, E.T. | | Deposit date: | 1998-06-10 | | Release date: | 1998-11-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Cation binding to the integrin CD11b I domain and activation model assessment
Structure, 6, 1998
|
|
3K2N
 
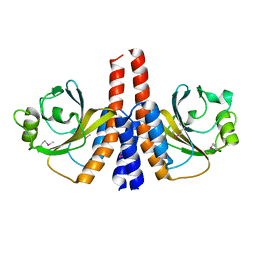 | |
3J6X
 
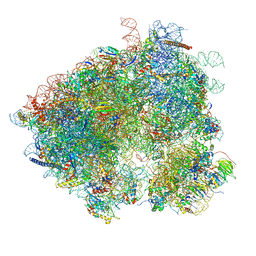 | | S. cerevisiae 80S ribosome bound with Taura syndrome virus (TSV) IRES, 5 degree rotation (Class II) | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 40S ribosomal protein S0, ... | | Authors: | Koh, C.S, Brilot, A.F, Grigorieff, N, Korostelev, A.A. | | Deposit date: | 2014-04-16 | | Release date: | 2014-06-11 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (6.1 Å) | | Cite: | Taura syndrome virus IRES initiates translation by binding its tRNA-mRNA-like structural element in the ribosomal decoding center.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
3J9Q
 
 | | Atomic structures of a bactericidal contractile nanotube in its pre- and post-contraction states | | Descriptor: | sheath, tube | | Authors: | Ge, P, Scholl, D, Leiman, P.G, Yu, X, Miller, J.F, Zhou, Z.H. | | Deposit date: | 2015-02-17 | | Release date: | 2015-04-01 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Atomic structures of a bactericidal contractile nanotube in its pre- and postcontraction states.
Nat.Struct.Mol.Biol., 22, 2015
|
|
3K35
 
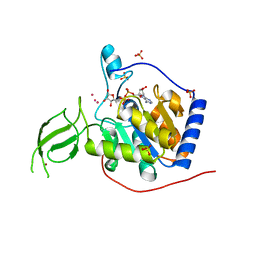 | | Crystal Structure of Human SIRT6 | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, NAD-dependent deacetylase sirtuin-6, SULFATE ION, ... | | Authors: | Pan, P.W, Dong, A, Qiu, W, Loppnau, P, Wang, J, Ravichandran, M, Bochkarev, A, Bountra, C, Weigelt, J, Arrowsmith, C.H, Min, J, Edwards, A.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-10-01 | | Release date: | 2009-12-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and biochemical functions of SIRT6.
J.Biol.Chem., 286, 2011
|
|
3J80
 
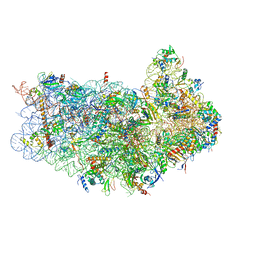 | | CryoEM structure of 40S-eIF1-eIF1A preinitiation complex | | Descriptor: | 18S rRNA, MAGNESIUM ION, RACK1, ... | | Authors: | Hussain, T, Llacer, J.L, Fernandez, I.S, Savva, C.G, Ramakrishnan, V. | | Deposit date: | 2014-08-28 | | Release date: | 2014-11-05 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.75 Å) | | Cite: | Structural changes enable start codon recognition by the eukaryotic translation initiation complex.
Cell(Cambridge,Mass.), 159, 2014
|
|
3JZ4
 
 | | Crystal structure of E. coli NADP dependent enzyme | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Succinate-semialdehyde dehydrogenase [NADP+] | | Authors: | Langendorf, C.G, Key, T.L.G, Fenalti, G, Kan, W.T, Buckle, A.M, Caradoc-Davies, T, Tuck, K.L, Law, R.H.P, Whisstock, J.C. | | Deposit date: | 2009-09-22 | | Release date: | 2010-03-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The X-ray crystal structure of Escherichia coli succinic semialdehyde dehydrogenase; structural insights into NADP+/enzyme interactions.
Plos One, 5, 2010
|
|
