7MCY
 
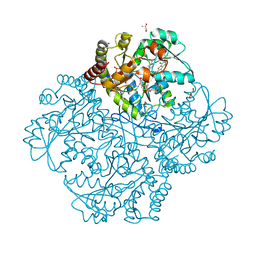 | | Crystal structure of Staphylococcus aureus Cystathionine gamma lyase, Holoenzyme with bound NL3 | | Descriptor: | 3-{[6-(7-chloro-1-benzothiophen-2-yl)-1H-indol-1-yl]methyl}-1H-pyrazole-5-carboxylic acid, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Bifunctional cystathionine gamma-lyase/homocysteine desulfhydrase, ... | | Authors: | Nuthanakanti, A, Serganov, A, Kaushik, A. | | Deposit date: | 2021-04-02 | | Release date: | 2021-06-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Inhibitors of bacterial H 2 S biogenesis targeting antibiotic resistance and tolerance.
Science, 372, 2021
|
|
7EPV
 
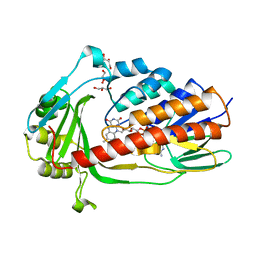 | | Crystal structure of tigecycline degrading monooxygenase Tet(X4) | | Descriptor: | DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, Flavin-dependent monooxygenase, GLYCEROL | | Authors: | Cheng, Q, Chen, S. | | Deposit date: | 2021-04-27 | | Release date: | 2021-11-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structural and mechanistic basis of the high catalytic activity of monooxygenase Tet(X4) on tigecycline.
Bmc Biol., 19, 2021
|
|
7MCU
 
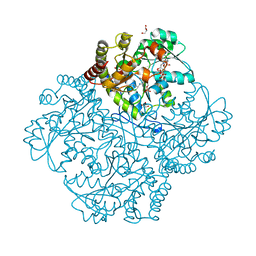 | | Crystal structure of Staphylococcus aureus Cystathionine gamma lyase, Holoenzyme with bound NL2 | | Descriptor: | 5-[(6-bromo-1H-indol-1-yl)methyl]-2-methylfuran-3-carboxylic acid, Bifunctional cystathionine gamma-lyase/homocysteine desulfhydrase, CITRATE ANION, ... | | Authors: | Nuthanakanti, A, Serganov, A, Kaushik, A. | | Deposit date: | 2021-04-02 | | Release date: | 2021-06-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Inhibitors of bacterial H 2 S biogenesis targeting antibiotic resistance and tolerance.
Science, 372, 2021
|
|
7EQZ
 
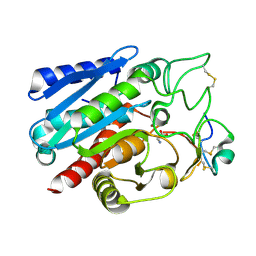 | | Crystal structure of Carboxypeptidase B complexed with Potato Carboxypeptidase Inhibitor | | Descriptor: | Carboxypeptidase B, GLYCINE, Metallocarboxypeptidase inhibitor, ... | | Authors: | Choong, Y.K, Gavor, E, Jobichen, C, Sivaraman, J. | | Deposit date: | 2021-05-05 | | Release date: | 2021-11-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of Aedes aegypti carboxypeptidase B1-inhibitor complex uncover the disparity between mosquito and non-mosquito insect carboxypeptidase inhibition mechanism.
Protein Sci., 30, 2021
|
|
2O24
 
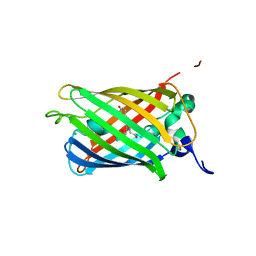 | |
7MD8
 
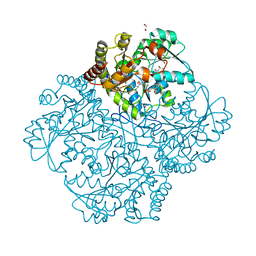 | | Crystal structure of Staphylococcus aureus cystathionine gamma lyase holoenzyme Y103N mutant co-crystallized with NL2 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Bifunctional cystathionine gamma-lyase/homocysteine desulfhydrase, GLYCEROL, ... | | Authors: | Nuthanakanti, A, Serganov, A, Kaushik, A. | | Deposit date: | 2021-04-03 | | Release date: | 2021-06-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Inhibitors of bacterial H 2 S biogenesis targeting antibiotic resistance and tolerance.
Science, 372, 2021
|
|
5N2F
 
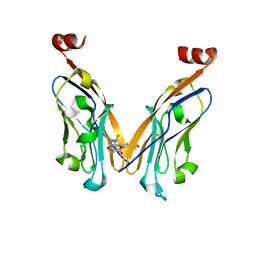 | | Structure of PD-L1/small-molecule inhibitor complex | | Descriptor: | 4-[[4-[[3-(2,3-dihydro-1,4-benzodioxin-6-yl)-2-methyl-phenyl]methoxy]-2,5-bis(fluoranyl)phenyl]methylamino]-3-oxidanylidene-butanoic acid, Programmed cell death 1 ligand 1 | | Authors: | Guzik, K, Zak, K.M, Grudnik, P, Dubin, G, Holak, T.A. | | Deposit date: | 2017-02-07 | | Release date: | 2017-06-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Small-Molecule Inhibitors of the Programmed Cell Death-1/Programmed Death-Ligand 1 (PD-1/PD-L1) Interaction via Transiently Induced Protein States and Dimerization of PD-L1.
J. Med. Chem., 60, 2017
|
|
2O29
 
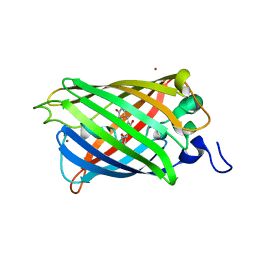 | |
2Z9X
 
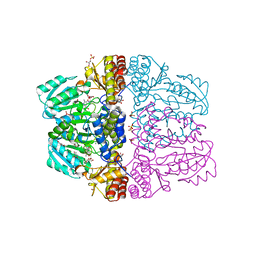 | | Crystal structure of pyridoxamine-pyruvate aminotransferase complexed with pyridoxyl-L-alanine | | Descriptor: | 3-HYDROXY-5-(HYDROXYMETHYL)-2-METHYLISONICOTINALDEHYDE, ALANINE, Aspartate aminotransferase, ... | | Authors: | Yoshikane, Y, Yokochi, N, Yamasaki, M, Mizutani, K, Ohnishi, K, Mikami, B, Hayashi, H, Yagi, T. | | Deposit date: | 2007-09-26 | | Release date: | 2007-11-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Crystal structure of pyridoxamine-pyruvate aminotransferase from Mesorhizobium loti MAFF303099
J.Biol.Chem., 283, 2008
|
|
7MDA
 
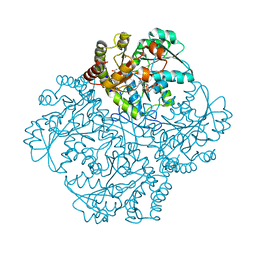 | | Crystal structure of Staphylococcus aureus cystathionine gamma lyase holoenzyme Y103A mutant co-crystallized with NL1 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Bifunctional cystathionine gamma-lyase/homocysteine desulfhydrase, N-[(6-bromo-1H-indol-1-yl)acetyl]glycine, ... | | Authors: | Nuthanakanti, A, Serganov, A, Kaushik, A. | | Deposit date: | 2021-04-03 | | Release date: | 2021-06-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Inhibitors of bacterial H 2 S biogenesis targeting antibiotic resistance and tolerance.
Science, 372, 2021
|
|
5N2N
 
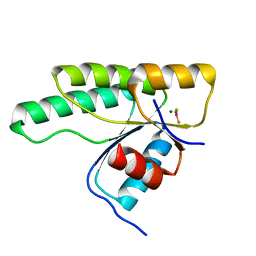 | | Crystal structure of the receiver domain of the histidine kinase CKI1 from Arabidopsis thaliana complexed with Mg2+ and BeF3- | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, Histidine kinase CKI1, MAGNESIUM ION | | Authors: | Otrusinova, O, Demo, G, Padrta, P, Jasenakova, Z, Pekarova, B, Gelova, Z, Szmitkowska, A, Kaderavek, P, Jansen, S, Zachrdla, M, Klumpler, T, Marek, J, Hritz, J, Janda, L, Iwai, H, Wimmerova, M, Hejatko, J, Zidek, L. | | Deposit date: | 2017-02-08 | | Release date: | 2017-09-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Conformational dynamics are a key factor in signaling mediated by the receiver domain of a sensor histidine kinase from Arabidopsis thaliana.
J. Biol. Chem., 292, 2017
|
|
2H3U
 
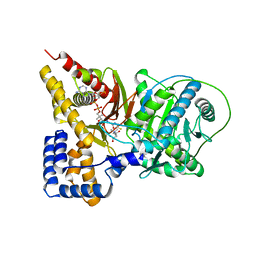 | |
5MZM
 
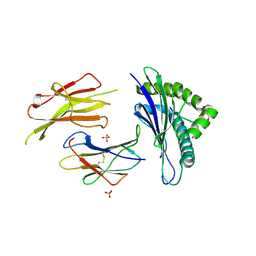 | | Structure of H-2Db in complex with TEIPP APL Trh4 p3P | | Descriptor: | Beta-2-microglobulin, Ceramide synthase 5 derived peptide Trh4 p3P, GLYCEROL, ... | | Authors: | Hafstrand, I, Achour, A, Sandalova, T. | | Deposit date: | 2017-02-01 | | Release date: | 2018-03-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The Immunogenicity of a Proline-Substituted Altered Peptide Ligand toward the Cancer-Associated TEIPP Neoepitope Trh4 Is Unrelated to Complex Stability.
J. Immunol., 200, 2018
|
|
2H5I
 
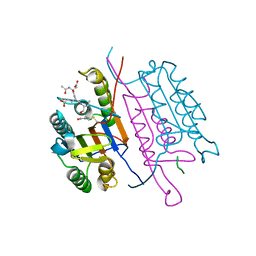 | | Crystal structure of caspase-3 with inhibitor Ac-DEVD-Cho | | Descriptor: | Ac-DEV(ASJ), caspase-3, p12 subunit, ... | | Authors: | Fang, B, Boross, P.I, Tozser, J, Weber, I.T. | | Deposit date: | 2006-05-26 | | Release date: | 2006-09-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Structural and kinetic analysis of caspase-3 reveals role for s5 binding site in substrate recognition
J.Mol.Biol., 360, 2006
|
|
2O3Z
 
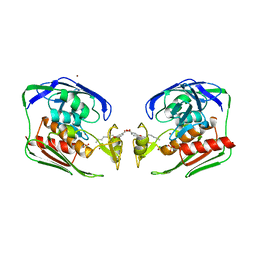 | | X-ray crystal structure of LpxC complexed with 3-heptyloxybenzoate | | Descriptor: | 3-(heptyloxy)benzoic acid, CHLORIDE ION, SULFATE ION, ... | | Authors: | Gennadios, H.A, Whittington, D.A, Christianson, D.W. | | Deposit date: | 2006-12-02 | | Release date: | 2007-02-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Amphipathic benzoic acid derivatives: synthesis and binding in the hydrophobic tunnel of the zinc deacetylase LpxC.
Bioorg.Med.Chem., 15, 2007
|
|
2H6N
 
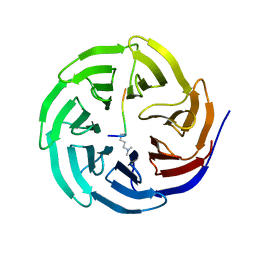 | | Histone H3 recognition and presentation by the WDR5 module of the MLL1 complex | | Descriptor: | Histone H3 K4-Me2 9-residue peptide, WD-repeat protein 5 | | Authors: | Ruthenburg, A.J, Wang, W.-K, Graybosch, D.M, Li, H, Allis, C.D, Patel, D.J, Verdine, G.L. | | Deposit date: | 2006-05-31 | | Release date: | 2006-07-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Histone H3 recognition and presentation by the WDR5 module of the MLL1 complex.
Nat.Struct.Mol.Biol., 13, 2006
|
|
5N3Q
 
 | | cAMP-dependent Protein Kinase A from Cricetulus griseus in complex with fragment like molecule 3-Aminobenzamide | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 3-aminobenzamide, DIMETHYL SULFOXIDE, ... | | Authors: | Siefker, C, Heine, A, Klebe, G. | | Deposit date: | 2017-02-08 | | Release date: | 2018-02-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Fragment Binding to Kinase Hinge: If Charge Distribution and Local pK a Shifts Mislead Popular Bioisosterism Concepts.
Angew.Chem.Int.Ed.Engl., 2020
|
|
2ZFD
 
 | | The crystal structure of plant specific calcium binding protein AtCBL2 in complex with the regulatory domain of AtCIPK14 | | Descriptor: | ACETIC ACID, CALCIUM ION, Calcineurin B-like protein 2, ... | | Authors: | Akaboshi, M, Hashimoto, H, Ishida, H, Koizumi, N, Sato, M, Shimizu, T. | | Deposit date: | 2007-12-29 | | Release date: | 2008-02-19 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | The crystal structure of plant-specific calcium-binding protein AtCBL2 in complex with the regulatory domain of AtCIPK14
J.Mol.Biol., 377, 2008
|
|
7MTB
 
 | | Rhodopsin kinase (GRK1)-S5E/S488E/T489E in complex with rhodopsin and Fab6 | | Descriptor: | Fab6 heavy chain, Fab6 light chain, RETINAL, ... | | Authors: | Chen, Q, Chen, C.-L, Tesmer, J.J.G. | | Deposit date: | 2021-05-13 | | Release date: | 2021-07-07 | | Last modified: | 2021-08-25 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structures of rhodopsin in complex with G-protein-coupled receptor kinase 1.
Nature, 595, 2021
|
|
5N1E
 
 | | cAMP-dependent Protein Kinase A from Cricetulus griseus in complex with fragment like molecule N-(1,3-benzodioxol-5-yl)-2-piperidin-1-ylacetamide | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, DIMETHYL SULFOXIDE, cAMP-dependent protein kinase catalytic subunit alpha, ... | | Authors: | Siefker, C, Heine, A, Klebe, G. | | Deposit date: | 2017-02-06 | | Release date: | 2018-02-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.529 Å) | | Cite: | A crystallographic fragment study with cAMP-dependent protein kinase A
To Be Published
|
|
2ZFQ
 
 | | Exploring thrombin S3 pocket | | Descriptor: | (S)-N-(4-carbamimidoylbenzyl)-1-(2-(cyclopentyloxy)ethanoyl)pyrrolidine-2-carboxamide, BENZAMIDINE, GLYCEROL, ... | | Authors: | Brandt, T, Baum, B, Heine, A, Klebe, G. | | Deposit date: | 2008-01-10 | | Release date: | 2009-01-13 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Exploring thrombin S3 pocket
To be Published
|
|
2ZG8
 
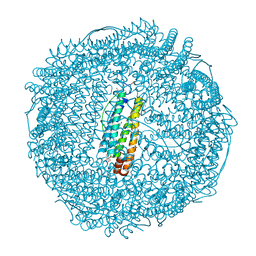 | | Crystal Structure of Pd(allyl)/apo-H49AFr | | Descriptor: | 1,2-ETHANEDIOL, CADMIUM ION, Ferritin light chain, ... | | Authors: | Abe, S, Niemeyer, J, Abe, M, Ueno, T, Hikage, T, Erker, G, Watanabe, Y. | | Deposit date: | 2008-01-18 | | Release date: | 2008-08-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Control of the coordination structure of organometallic palladium complexes in an apo-ferritin cage.
J.Am.Chem.Soc., 130, 2008
|
|
7M60
 
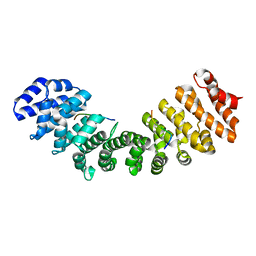 | | Structure of Mouse Importin alpha MLH1-S467A NLS Peptide Complex | | Descriptor: | DNA mismatch repair protein Mlh1 NLS peptide, Importin subunit alpha-1 | | Authors: | De Oliveira, H.C, Da Silva, T.D, Fukuda, C.A, Fontes, M.R.M. | | Deposit date: | 2021-03-25 | | Release date: | 2021-07-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and calorimetric studies reveal specific determinants for the binding of a high-affinity NLS to mammalian importin-alpha.
Biochem.J., 478, 2021
|
|
3KAT
 
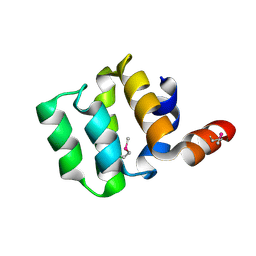 | | Crystal Structure of the CARD domain of the human NLRP1 protein, Northeast Structural Genomics Consortium Target HR3486E | | Descriptor: | NACHT, LRR and PYD domains-containing protein 1 | | Authors: | Forouhar, F, Abashidze, M, Seetharaman, J, Mao, M, Xiao, R, Ciccosanti, C, Shastry, R, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-10-19 | | Release date: | 2009-10-27 | | Last modified: | 2019-07-17 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Northeast Structural Genomics Consortium Target HR3486E
To be Published
|
|
5N52
 
 | |
