1WAW
 
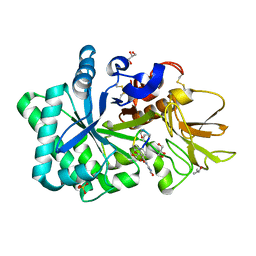 | | Specificity and affinity of natural product cyclopentapeptide inhibitor Argadin against human chitinase | | Descriptor: | ARGADIN, CHITOTRIOSIDASE 1, GLYCEROL, ... | | Authors: | Rao, F.V, Houston, D.R, Boot, R.G, Aerts, J.M.F.G, Hodkinson, M, Adams, D.J, Shiomi, K, Omura, S, van Aalten, D.M.F. | | Deposit date: | 2004-10-28 | | Release date: | 2005-01-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Specificity and Affinity of Natural Product Cyclopentapeptide Inhibitors Against Aspergillus Fumigatus, Human and Bacterial Chitinases
Chem.Biol., 12, 2005
|
|
5WF0
 
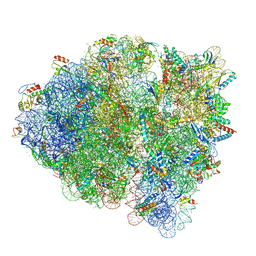 | |
1VYF
 
 | | schistosoma mansoni fatty acid binding protein in complex with oleic acid | | Descriptor: | 14 KDA FATTY ACID BINDING PROTEIN, OLEIC ACID | | Authors: | Angelucci, F, Johnson, K.A, Baiocco, P, Miele, A.E, Bellelli, A. | | Deposit date: | 2004-04-29 | | Release date: | 2004-09-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Schistosoma Mansoni Fatty Acid Binding Protein: Specificity and Functional Control as Revealed by Crystallographic Structure
Biochemistry, 43, 2004
|
|
5WSZ
 
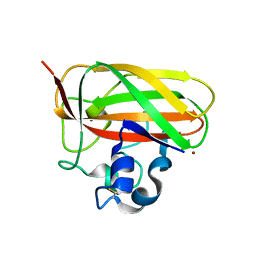 | | Crystal structure of a lytic polysaccharide monooxygenase,BtLPMO10A, from Bacillus thuringiensis | | Descriptor: | COPPER (II) ION, LpmO10A | | Authors: | Zhao, Y, Zhang, H, Yin, H. | | Deposit date: | 2016-12-08 | | Release date: | 2017-02-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.565 Å) | | Cite: | Crystal structure of a lytic polysaccharide monooxygenase,BtLPMO10A, from Bacillus thuringiensis
To Be Published
|
|
1VZ2
 
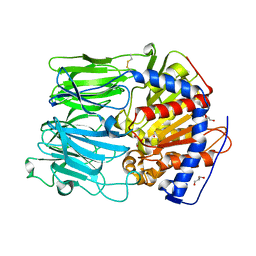 | |
5WTT
 
 | | Structure of the 093G9 Fab in complex with the epitope peptide | | Descriptor: | Epitope peptide of Cyr61, Heavy chain of 093G9 Fab, Light chain of 093G9 Fab | | Authors: | Zhong, C, Hu, K, Shen, J, Ding, J. | | Deposit date: | 2016-12-14 | | Release date: | 2017-12-20 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Molecular basis for the recognition of CCN1 by monoclonal antibody 093G9.
J. Mol. Recognit., 30, 2017
|
|
1W40
 
 | | T. celer L30e K9A variant | | Descriptor: | 50S RIBOSOMAL PROTEIN L30E | | Authors: | Lee, C.F, Lee, K.M, Chan, S.H, Allen, M.D, Bycroft, M, Wong, K.B. | | Deposit date: | 2004-07-22 | | Release date: | 2005-04-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Electrostatic Interactions Contribute to Reduced Heat Capacity Change of Unfolding in a Thermophilic Ribosomal Protein L30E
J.Mol.Biol., 348, 2005
|
|
1VZW
 
 | | Crystal structure of the bifunctional protein Pria | | Descriptor: | GLYCEROL, PHOSPHORIBOSYL ISOMERASE A, SULFATE ION | | Authors: | Kuper, J, Wilmanns, M. | | Deposit date: | 2004-05-27 | | Release date: | 2005-01-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Two-Fold Repeated (Beta-Alpha)(4) Half-Barrels May Provide a Molecular Tool for Dual Substrate Specificity
Embo Rep., 6, 2005
|
|
5WUW
 
 | |
1W5B
 
 | | FtsZ dimer, GTP soak (M. jannaschii) | | Descriptor: | CELL DIVISION PROTEIN FTSZ HOMOLOG 1, GUANOSINE-5'-TRIPHOSPHATE | | Authors: | Oliva, M.A, Cordell, S.C, Lowe, J. | | Deposit date: | 2004-08-06 | | Release date: | 2004-12-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Insights Into Ftsz Protofilament Formation
Nat.Struct.Mol.Biol., 11, 2004
|
|
5W7I
 
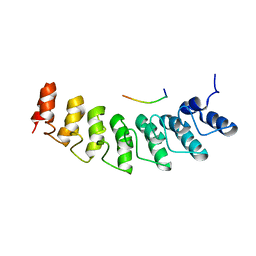 | | X-ray structure of ankyrin repeat domain of DHHC17 in complex with Snap25b peptide | | Descriptor: | Palmitoyltransferase ZDHHC17, Snap25b-111-120 | | Authors: | Verardi, R, Kim, J.-S, Ghirlando, R, Banerjee, A. | | Deposit date: | 2017-06-20 | | Release date: | 2017-08-09 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.105 Å) | | Cite: | Structural Basis for Substrate Recognition by the Ankyrin Repeat Domain of Human DHHC17 Palmitoyltransferase.
Structure, 25, 2017
|
|
1W2G
 
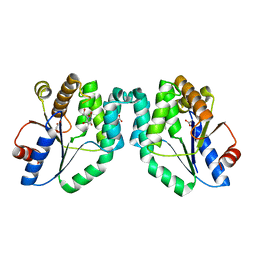 | | Crystal Structure Of Mycobacterium Tuberculosis Thymidylate Kinase Complexed With Deoxythymidine (dT) (2.1 A Resolution) | | Descriptor: | ACETATE ION, THYMIDINE, THYMIDYLATE KINASE TMK | | Authors: | Fioravanti, E, Adam, V, Munier-Lehmann, H, Bourgeois, D. | | Deposit date: | 2004-07-06 | | Release date: | 2005-01-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Crystal Structure of Mycobacterium Tuberculosis Thymidylate Kinase in Complex with 3'-Azidodeoxythymidine Monophosphate Suggests a Mechanism for Competitive Inhibition
Biochemistry, 44, 2005
|
|
1W1L
 
 | |
1W15
 
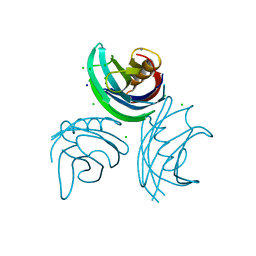 | | rat synaptotagmin 4 C2B domain in the presence of calcium | | Descriptor: | CALCIUM ION, CHLORIDE ION, SODIUM ION, ... | | Authors: | Dai, H, Shin, O.-H, Machius, M, Tomchick, D.R, Sudhof, T.C, Rizo, J. | | Deposit date: | 2004-06-16 | | Release date: | 2004-08-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Structural Basis for the Evolutionary Inactivation of Ca2+ Binding to Synaptotagmin 4
Nat.Struct.Mol.Biol., 11, 2004
|
|
1W2H
 
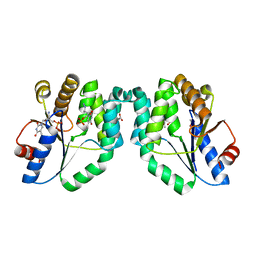 | | Crystal Structure Of Mycobacterium Tuberculosis Thymidylate Kinase Complexed With Azidothymidine Monophosphate (AZT-MP) (2.0 A Resolution) | | Descriptor: | 3'-AZIDO-3'-DEOXYTHYMIDINE-5'-MONOPHOSPHATE, ACETATE ION, THYMIDYLATE KINASE TMK | | Authors: | Fioravanti, E, Adam, V, Munier-Lehmann, H, Bourgeois, D. | | Deposit date: | 2004-07-06 | | Release date: | 2005-01-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of Mycobacterium tuberculosis thymidylate kinase in complex with 3'-azidodeoxythymidine monophosphate suggests a mechanism for competitive inhibition.
Biochemistry, 44, 2005
|
|
1W5M
 
 | | Stepwise introduction of zinc binding site into porphobilinogen synthase of Pseudomonas aeruginosa (mutations A129C and D139C) | | Descriptor: | CHLORIDE ION, DELTA-AMINOLEVULINIC ACID DEHYDRATASE, FORMIC ACID, ... | | Authors: | Frere, F, Reents, H, Schubert, W.-D, Heinz, D.W, Jahn, D. | | Deposit date: | 2004-08-09 | | Release date: | 2005-01-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Tracking the Evolution of Porphobilinogen Synthase Metal Dependence in Vitro
J.Mol.Biol., 345, 2005
|
|
1W50
 
 | | Apo Structure of BACE (Beta Secretase) | | Descriptor: | BETA-SECRETASE 1, IODIDE ION | | Authors: | Patel, S, Vuillard, L, Cleasby, A, Murray, C.W, Yon, J. | | Deposit date: | 2004-08-04 | | Release date: | 2004-09-23 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Apo and Inhibitor Complex Structures of Bace (Beta-Secretase)
J.Mol.Biol., 343, 2004
|
|
1W6Q
 
 | | X-RAY CRYSTAL STRUCTURE OF R111H HUMAN GALECTIN-1 | | Descriptor: | BETA-MERCAPTOETHANOL, GALECTIN-1 | | Authors: | Lopez-Lucendo, M.I.F, Gabius, H.J, Romero, A. | | Deposit date: | 2004-08-21 | | Release date: | 2004-10-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Growth-Regulatory Human Galectin-1: Crystallographic Characterisation of the Structural Changes Induced by Single-Site Mutations and Their Impact on the Thermodynamics of Ligand Binding
J.Mol.Biol., 343, 2004
|
|
5LUB
 
 | | Crystal structure of human legumain (AEP) in complex with compound 11 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 7-(morpholin-4-yl)-2,1,3-benzoxadiazol-4-amine, ... | | Authors: | Dall, E, Ye, K, Brandstetter, H. | | Deposit date: | 2016-09-08 | | Release date: | 2017-03-29 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Inhibition of delta-secretase improves cognitive functions in mouse models of Alzheimer's disease.
Nat Commun, 8, 2017
|
|
1VYP
 
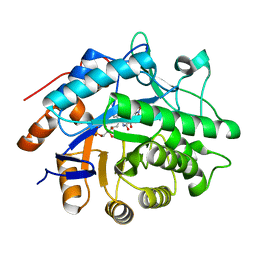 | | Structure of pentaerythritol tetranitrate reductase W102F mutant and complexed with picric acid | | Descriptor: | FLAVIN MONONUCLEOTIDE, PENTAERYTHRITOL TETRANITRATE REDUCTASE, PICRIC ACID | | Authors: | Barna, T, Moody, P.C.E. | | Deposit date: | 2004-05-04 | | Release date: | 2004-05-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | Atomic Resolution Structures and Solution Behavior of Enzyme-Substrate Complexes of Enterobacter Cloacae Pb2 Pentaerythritol Tetranitrate Reductase: Multiple Conformational States and Implications for the Mechanism of Nitroaromatic Explosive Degradation
J.Biol.Chem., 279, 2004
|
|
7OAN
 
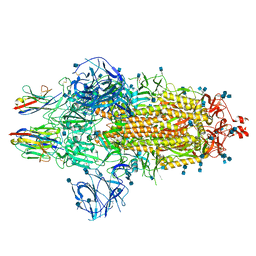 | | Nanobody C5 bound to Spike | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Naismith, J.H, Weckener, M. | | Deposit date: | 2021-04-19 | | Release date: | 2021-08-11 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | A potent SARS-CoV-2 neutralising nanobody shows therapeutic efficacy in the Syrian golden hamster model of COVID-19.
Nat Commun, 12, 2021
|
|
1VZO
 
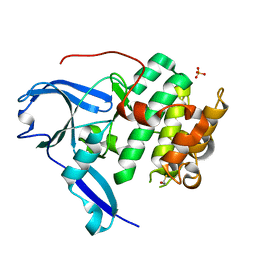 | | The structure of the N-terminal kinase domain of MSK1 reveals a novel autoinhibitory conformation for a dual kinase protein | | Descriptor: | BETA-MERCAPTOETHANOL, RIBOSOMAL PROTEIN S6 KINASE ALPHA 5, SULFATE ION | | Authors: | Smith, K.J, Carter, P.S, Bridges, A, Horrocks, P, Lewis, C, Pettman, G, Clarke, A, Brown, M, Hughes, J, Wilkinson, M, Bax, B, Reith, A. | | Deposit date: | 2004-05-21 | | Release date: | 2004-06-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Structure of Msk1 Reveals a Novel Autoinhibitory Conformation for a Dual Kinase Protein
Structure, 12, 2004
|
|
1W3C
 
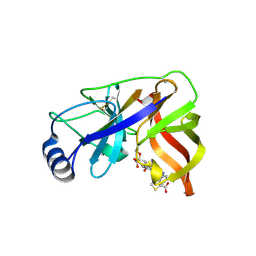 | | Crystal structure of the Hepatitis C Virus NS3 Protease in complex with a peptidomimetic inhibitor | | Descriptor: | 3-({(2S)-2-[({(1R)-1-[({(1R)-1-[(R)-CARBOXY(HYDROXY)METHYL]-3,3-DIFLUOROPROPYL}AMINO)CARBONYL]-3-METHYLBUTYL}AMINO)CARB ONYL]-2,3-DIHYDRO-1H-INDOL-2-YL}METHYL)THIOPHENE-2-CARBOXYLIC ACID, 3-({(2S)-2-[({(1S)-1-[({(1S)-1-[(R)-CARBOXY(HYDROXY)METHYL]-3,3-DIFLUOROPROPYL}AMINO)CARBONYL]-3-METHYLBUTYL}AMINO)CARBONYL]-2,3-DIHYDRO-1H-INDOL-2-YL}METHYL)THIOPHENE-2-CARBOXYLIC ACID, NONSTRUCTURAL PROTEIN NS4A (P4), ... | | Authors: | Di Marco, S, Volpari, C. | | Deposit date: | 2004-07-14 | | Release date: | 2004-12-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Design and Enzyme-Bound Crystal Structure of Indoline Based Peptidomimetic Inhibitors of Hepatitis C Virus Ns3 Protease
J.Med.Chem., 47, 2004
|
|
1W19
 
 | | Lumazine Synthase from Mycobacterium tuberculosis bound to 3-(1,3,7- trihydro-9-D-ribityl-2,6,8-purinetrione-7-yl)propane 1-phosphate | | Descriptor: | (2R,3S)-1,4-DIMERCAPTOBUTANE-2,3-DIOL, (2S,3S)-1,4-DIMERCAPTOBUTANE-2,3-DIOL, (4S,5S)-1,2-DITHIANE-4,5-DIOL, ... | | Authors: | Morgunova, E, Meining, W, Illarionov, B, Haase, I, Fischer, M, Cushman, M, Bacher, A, Ladenstein, R. | | Deposit date: | 2004-06-03 | | Release date: | 2005-03-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Lumazine Synthase from Mycobacterium Tuberculosis as a Target for Rational Drug Design: Binding Mode of a New Class of Purinetrione Inhibitors(,)
Biochemistry, 44, 2005
|
|
1W23
 
 | | Crystal structure of phosphoserine aminotransferase from Bacillus alcalophilus | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Dubnovitsky, A, Kapetaniou, E.G, Papageorgiou, A.C. | | Deposit date: | 2004-06-25 | | Release date: | 2004-12-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Enzyme Adaptation to Alkaline Ph: Atomic Resolution (1.08 A) Structure of Phosphoserine Aminotransferase from Bacillus Alcalophilus
Protein Sci., 14, 2005
|
|
