3QB8
 
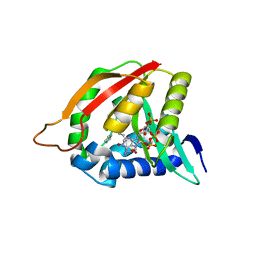 | | Paramecium Chlorella Bursaria Virus1 Putative ORF A654L is a Polyamine Acetyltransferase | | Descriptor: | A654L protein, COENZYME A, IMIDAZOLE | | Authors: | Charlop-Powers, Z, Zhou, M.-M, Jakoncic, J, Gurnon, J, Van Etten, J. | | Deposit date: | 2011-01-12 | | Release date: | 2012-01-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Paramecium bursaria chlorella virus 1 encodes a polyamine acetyltransferase.
J. Biol. Chem., 287, 2012
|
|
5R89
 
 | |
4BQ4
 
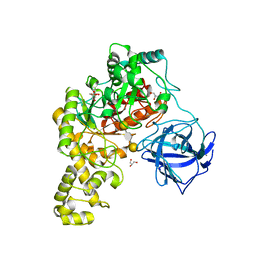 | | Structural analysis of an exo-beta-agarase | | Descriptor: | 3,6-anhydro-alpha-L-galactopyranose-(1-3)-beta-D-galactopyranose-(1-4)-3,6-anhydro-alpha-L-galactopyranose-(1-3)-beta-D-galactopyranose, B-AGARASE, CALCIUM ION, ... | | Authors: | Pluvinage, B, Hehemann, J.H, Boraston, A.B. | | Deposit date: | 2013-05-29 | | Release date: | 2013-08-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Substrate Recognition and Hydrolysis by a Family 50 Exo-Beta-Agarase Aga50D from the Marine Bacterium Saccharophagus Degradans
J.Biol.Chem., 288, 2013
|
|
2GIQ
 
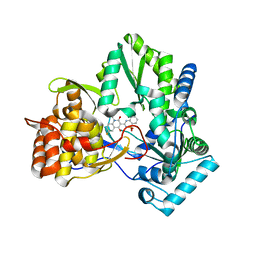 | | Hepatitis C virus RNA-dependent RNA polymerase NS5B with NNI-2 inhibitor | | Descriptor: | 1-(2-CYCLOPROPYLETHYL)-3-(1,1-DIOXIDO-2H-1,2,4-BENZOTHIADIAZIN-3-YL)-6-FLUORO-4-HYDROXYQUINOLIN-2(1H)-ONE, RNA-directed RNA polymerase | | Authors: | Harris, S.F. | | Deposit date: | 2006-03-29 | | Release date: | 2007-04-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Selection and characterization of replicon variants dually resistant to thumb- and palm-binding nonnucleoside polymerase inhibitors of the hepatitis C virus.
J.Virol., 80, 2006
|
|
3QLR
 
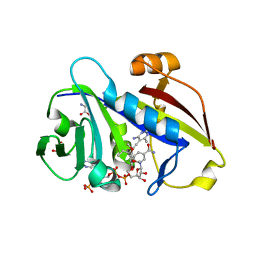 | | Candida albicans dihydrofolate reductase complexed with NADPH and 6-methyl-5-[(3R)-3-(3,4,5-trimethoxyphenyl)pent-1-yn-1-yl]pyrimidine-2,4-diamine (UCP112A) | | Descriptor: | 6-methyl-5-[(3R)-3-(3,4,5-trimethoxyphenyl)pent-1-yn-1-yl]pyrimidine-2,4-diamine, GLYCEROL, GLYCINE, ... | | Authors: | Paulsen, J.L, Bendel, S.D, Anderson, A.C. | | Deposit date: | 2011-02-03 | | Release date: | 2011-07-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.149 Å) | | Cite: | Crystal Structures of Candida albicans Dihydrofolate Reductase Bound to Propargyl-Linked Antifolates Reveal the Flexibility of Active Site Loop Residues Critical for Ligand Potency and Selectivity.
Chem.Biol.Drug Des., 78, 2011
|
|
3QKV
 
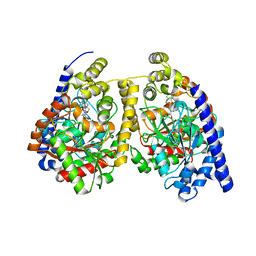 | | Crystal structure of fatty acid amide hydrolase with small molecule compound | | Descriptor: | (6-bromo-1'H,4H-spiro[1,3-benzodioxine-2,4'-piperidin]-1'-yl)methanol, Fatty-acid amide hydrolase 1 | | Authors: | Min, X, Walker, N.P.C, Wang, Z. | | Deposit date: | 2011-02-01 | | Release date: | 2011-04-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Discovery and molecular basis of potent noncovalent inhibitors of fatty acid amide hydrolase (FAAH).
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3AIS
 
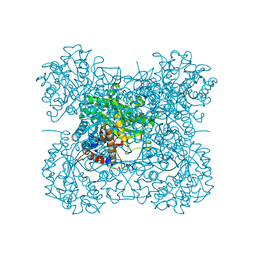 | | Crystal structure of a mutant beta-glucosidase in wheat complexed with DIMBOA-Glc | | Descriptor: | (2S)-2,4-dihydroxy-7-methoxy-2H-1,4-benzoxazin-3(4H)-one, Beta-glucosidase, beta-D-glucopyranose | | Authors: | Sue, M, Nakamura, C, Miyamoto, T, Yajima, S. | | Deposit date: | 2010-05-18 | | Release date: | 2011-03-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Active-site architecture of benzoxazinone-glucoside beta-D-glucosidases in Triticeae
Plant Sci., 180, 2011
|
|
5VTW
 
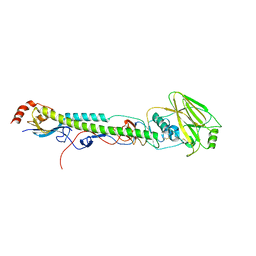 | |
7FQH
 
 | | Crystal Structure of human Legumain in complex with (2S)-N-[(3S)-5-amino-5-oxopent-1-yn-3-yl]-1-[1-[4-(cyclopropylmethoxy)phenyl]cyclopropanecarbonyl]pyrrolidine-2-carboxamide | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Legumain, ... | | Authors: | Ehler, A, Benz, J, Bartels, B, Hewings-David, S, Rudolph, M.G. | | Deposit date: | 2022-10-05 | | Release date: | 2023-10-11 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Crystal Structure of a human Legumain complex
To be published
|
|
4IMD
 
 | |
3R88
 
 | | Anthranilate phosphoribosyltransferase (trpD) from Mycobacterium tuberculosis (complex with inhibitor ACS145) | | Descriptor: | 1-O-pyrophosphono-5-O-phosphono-alpha-D-ribofuranose, 2-amino-4,5-dimethoxybenzoic acid, Anthranilate phosphoribosyltransferase, ... | | Authors: | Castell, A, Short, F.L, Lott, J.S. | | Deposit date: | 2011-03-23 | | Release date: | 2012-09-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | The Substrate Capture Mechanism of Mycobacterium tuberculosis Anthranilate Phosphoribosyltransferase Provides a Mode for Inhibition.
Biochemistry, 52, 2013
|
|
1Q2U
 
 | | Crystal structure of DJ-1/RS and implication on familial Parkinson's disease | | Descriptor: | RNA-binding protein regulatory subunit | | Authors: | Huai, Q, Sun, Y, Wang, H, Chin, L.S, Li, L, Robinson, H, Ke, H. | | Deposit date: | 2003-07-26 | | Release date: | 2003-10-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of DJ-1/RS and implication on familial Parkinson's disease
Febs Lett., 549, 2003
|
|
3V2K
 
 | | Crystal structure of ribosome inactivating protein from momordica balsamina complexed with the product of RNA substrate adenosine triphosphate at 2.0 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ADENINE, GLYCEROL, ... | | Authors: | Kushwaha, G.S, Pandey, N, Sinha, M, Bhushan, A, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2011-12-12 | | Release date: | 2012-01-18 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Crystal structures of a type-1 ribosome inactivating protein from Momordica balsamina in the bound and unbound states.
Biochim.Biophys.Acta, 1824, 2012
|
|
1UBZ
 
 | |
3EOB
 
 | |
3OE8
 
 | | Crystal structure of the CXCR4 chemokine receptor in complex with a small molecule antagonist IT1t in P1 spacegroup | | Descriptor: | (6,6-dimethyl-5,6-dihydroimidazo[2,1-b][1,3]thiazol-3-yl)methyl N,N'-dicyclohexylimidothiocarbamate, C-X-C chemokine receptor type 4, Lysozyme Chimera | | Authors: | Wu, B, Mol, C.D, Han, G.W, Katritch, V, Chien, E.Y.T, Liu, W, Cherezov, V, Stevens, R.C, Accelerated Technologies Center for Gene to 3D Structure (ATCG3D), GPCR Network (GPCR) | | Deposit date: | 2010-08-12 | | Release date: | 2010-10-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structures of the CXCR4 chemokine GPCR with small-molecule and cyclic peptide antagonists.
Science, 330, 2010
|
|
1GFI
 
 | | STRUCTURES OF ACTIVE CONFORMATIONS OF GI ALPHA 1 AND THE MECHANISM OF GTP HYDROLYSIS | | Descriptor: | GUANINE NUCLEOTIDE-BINDING PROTEIN G, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Coleman, D.E, Berghuis, A.M, Sprang, S.R. | | Deposit date: | 1994-11-11 | | Release date: | 1995-03-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structures of active conformations of Gi alpha 1 and the mechanism of GTP hydrolysis.
Science, 265, 1994
|
|
4OFD
 
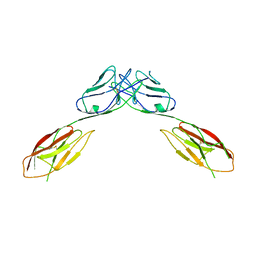 | | Crystal Structure of mouse Neph1 D1-D2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Kin of IRRE-like protein 1, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Ozkan, E, Garcia, K.C. | | Deposit date: | 2014-01-14 | | Release date: | 2014-02-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.94 Å) | | Cite: | Extracellular Architecture of the SYG-1/SYG-2 Adhesion Complex Instructs Synaptogenesis.
Cell(Cambridge,Mass.), 156, 2014
|
|
1GJF
 
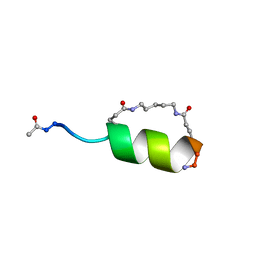 | | Peptide Antagonist of IGFBP1, (i,i+7) Covalently Restrained Analog, Minimized Average Structure | | Descriptor: | IGFBP-1 antagonist, PENTANE | | Authors: | Skelton, N.J, Chen, Y.M, Dubree, N, Quan, C, Jackson, D.Y, Cochran, A.G, Zobel, K, Deshayes, K, Baca, M, Pisabarro, M.T, Lowman, H.B. | | Deposit date: | 2001-05-11 | | Release date: | 2001-05-30 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Structure-function analysis of a phage display-derived peptide that binds to insulin-like growth factor binding protein 1.
Biochemistry, 40, 2001
|
|
3OKU
 
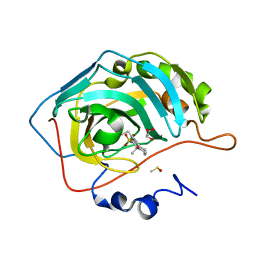 | | Human Carbonic Anhydrase II in complex with 2-Ethylestrone-3-O-sulfamate | | Descriptor: | (9beta)-2-ethyl-17-oxoestra-1(10),2,4-trien-3-yl sulfamate, Carbonic anhydrase 2, DIMETHYL SULFOXIDE, ... | | Authors: | Sippel, K.H, Stander, B.A, Robbins, A.H, Tu, C.K, Agbandje-McKenna, M, Silverman, D.N, Joubert, A.M, McKenna, R. | | Deposit date: | 2010-08-25 | | Release date: | 2011-07-06 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Characterization of Carbonic Anhydrase Isozyme Specific Inhibition by Sulfamated 2-Ethylestra Compounds
LETT.DRUG DES.DISCOVERY, 8, 2011
|
|
3OIM
 
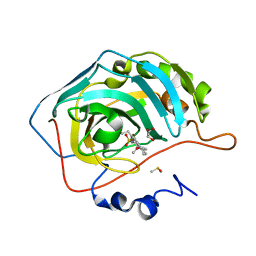 | | Human Carbonic anhydrase II bound by 2-Ethylestradiol 3-O-sulfamate | | Descriptor: | (14beta,17alpha)-2-ethyl-17-hydroxyestra-1(10),2,4-trien-3-yl sulfamate, Carbonic anhydrase 2, DIMETHYL SULFOXIDE, ... | | Authors: | Sippel, K.H, Stander, B.A, Robbins, A.H, Tu, C.K, Agbandje-McKenna, M, Silverman, D.N, Joubert, A.M, McKenna, R. | | Deposit date: | 2010-08-19 | | Release date: | 2011-07-06 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Characterization of Carbonic Anhydrase Isozyme Specific Inhibition by Sulfamated 2-Ethylestra Compounds
LETT.DRUG DES.DISCOVERY, 8, 2011
|
|
4OI9
 
 | | Crystal Structure of ICAM-5 D1-D4 ectodomain fragment, Space Group P21 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Recacha, R, Jimenez, D, Tian, L, Barredo, R, Ghamberg, C, Casasnovas, J.M. | | Deposit date: | 2014-01-19 | | Release date: | 2014-07-16 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of an ICAM-5 ectodomain fragment show electrostatic-based homophilic adhesions.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
1ZZL
 
 | | Crystal structure of P38 with triazolopyridine | | Descriptor: | 6-[4-(4-FLUOROPHENYL)-1,3-OXAZOL-5-YL]-3-ISOPROPYL[1,2,4]TRIAZOLO[4,3-A]PYRIDINE, Mitogen-activated protein kinase 14 | | Authors: | McClure, K.F, Han, S. | | Deposit date: | 2005-06-14 | | Release date: | 2005-09-13 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Theoretical and Experimental Design of Atypical Kinase Inhibitors: Application to p38 MAP Kinase.
J.Med.Chem., 48, 2005
|
|
3OLF
 
 | |
1FXJ
 
 | | CRYSTAL STRUCTURE OF N-ACETYLGLUCOSAMINE 1-PHOSPHATE URIDYLTRANSFERASE | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, SULFATE ION, UDP-N-ACETYLGLUCOSAMINE PYROPHOSPHORYLASE | | Authors: | Brown, K, Pompeo, F, Dixon, S, Mengin-Lecreulx, D, Cambillau, C, Bourne, Y. | | Deposit date: | 2000-09-26 | | Release date: | 2000-10-18 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structure of the bifunctional N-acetylglucosamine 1-phosphate uridyltransferase from Escherichia coli: a paradigm for the related pyrophosphorylase superfamily.
EMBO J., 18, 1999
|
|
