8E6W
 
 | | Escherichia coli Rho-dependent transcription pre-termination complex containing 18 nt long RNA spacer, lambda-tR1 rut RNA, Mg-ADP-BeF3, and NusG; Rho part | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, MAGNESIUM ION, ... | | Authors: | Molodtsov, V, Wang, C, Ebright, R.H. | | Deposit date: | 2022-08-23 | | Release date: | 2022-09-07 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (4.27 Å) | | Cite: | Structural basis of Rho-dependent transcription termination.
Nature, 614, 2023
|
|
8E5P
 
 | | Escherichia coli Rho-dependent transcription pre-termination complex containing 24 nt long RNA spacer, Mg-ADP-BeF3, and NusG; Rho hexamer part | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, MAGNESIUM ION, ... | | Authors: | Molodtsov, V, Wang, C. | | Deposit date: | 2022-08-22 | | Release date: | 2022-09-07 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Structural basis of Rho-dependent transcription termination.
Nature, 614, 2023
|
|
6UKA
 
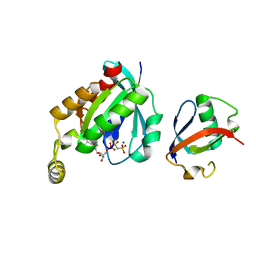 | | Crystal structure of RHOG and ELMO complex | | Descriptor: | Engulfment and cell motility protein 2, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, ... | | Authors: | Jo, C.H, Killoran, R.C, Smith, M.J. | | Deposit date: | 2019-10-04 | | Release date: | 2020-08-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of the DOCK2-ELMO1 complex provides insights into regulation of the auto-inhibited state.
Nat Commun, 11, 2020
|
|
6UN0
 
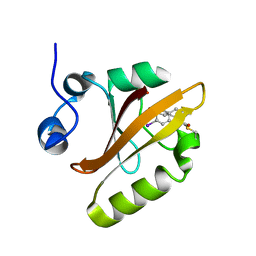 | |
4QA3
 
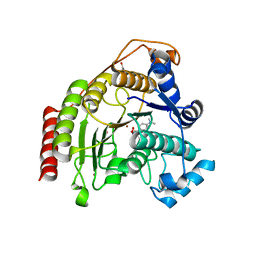 | | Crystal structure of T311M HDAC8 in complex with Trichostatin A (TSA) | | Descriptor: | GLYCEROL, Histone deacetylase 8, POTASSIUM ION, ... | | Authors: | Decroos, C, Bowman, C.B, Moser, J.-A.S, Christianson, K.E, Deardorff, M.A, Christianson, D.W. | | Deposit date: | 2014-05-02 | | Release date: | 2014-08-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.876 Å) | | Cite: | Compromised Structure and Function of HDAC8 Mutants Identified in Cornelia de Lange Syndrome Spectrum Disorders.
Acs Chem.Biol., 9, 2014
|
|
6UNF
 
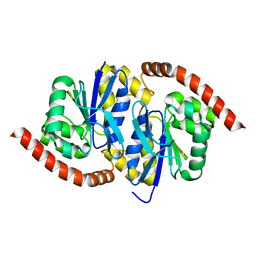 | |
6CEW
 
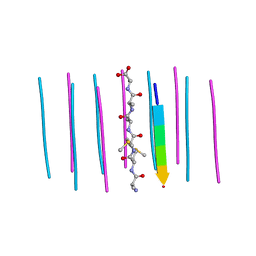 | | Segment AMMAAA from the low complexity domain of TDP-43, residues 321-326 | | Descriptor: | AMMAAA | | Authors: | Guenther, E.L, Cao, Q, Lu, J, Sawaya, M.R, Eisenberg, D.S. | | Deposit date: | 2018-02-12 | | Release date: | 2018-04-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Atomic structures of TDP-43 LCD segments and insights into reversible or pathogenic aggregation.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6CF4
 
 | | Segment NFGTFS, with familial mutation A315T and phosphorylated threonine, from the low complexity domain of TDP-43, residues 312-317 | | Descriptor: | NFGTFS | | Authors: | Guenther, E.L, Cao, Q, Boyer, D.R, Sawaya, M.R, Eisenberg, D.S. | | Deposit date: | 2018-02-13 | | Release date: | 2018-05-23 | | Last modified: | 2021-06-30 | | Method: | ELECTRON CRYSTALLOGRAPHY (0.75 Å) | | Cite: | Atomic structures of TDP-43 LCD segments and insights into reversible or pathogenic aggregation.
Nat. Struct. Mol. Biol., 25, 2018
|
|
4QA0
 
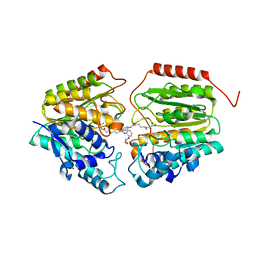 | | Crystal structure of C153F HDAC8 in complex with SAHA | | Descriptor: | GLYCEROL, Histone deacetylase 8, OCTANEDIOIC ACID HYDROXYAMIDE PHENYLAMIDE, ... | | Authors: | Decroos, C, Bowman, C.B, Moser, J.-A.S, Christianson, K.E, Deardorff, M.A, Christianson, D.W. | | Deposit date: | 2014-05-02 | | Release date: | 2014-08-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.242 Å) | | Cite: | Compromised Structure and Function of HDAC8 Mutants Identified in Cornelia de Lange Syndrome Spectrum Disorders.
Acs Chem.Biol., 9, 2014
|
|
2VV7
 
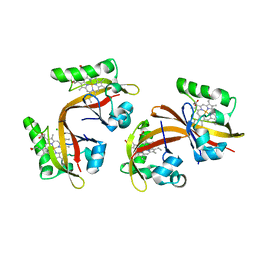 | | BJFIXLH IN UNLIGANDED FERROUS FORM | | Descriptor: | CHLORIDE ION, PROTOPORPHYRIN IX CONTAINING FE, SENSOR PROTEIN FIXL, ... | | Authors: | Ayers, R.A, Moffat, K. | | Deposit date: | 2008-06-04 | | Release date: | 2008-11-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Changes in Quaternary Structure in the Signaling Mechanisms of Pas Domains.
Biochemistry, 47, 2008
|
|
2Y48
 
 | | Crystal structure of LSD1-CoREST in complex with a N-terminal SNAIL peptide | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, LYSINE-SPECIFIC DEMETHYLASE 1A, REST COREPRESSOR 1, ... | | Authors: | Baron, R, Binda, C, Tortorici, M, McCammon, J.A, Mattevi, A. | | Deposit date: | 2011-01-05 | | Release date: | 2011-02-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Molecular Mimicry and Ligand Recognition in Binding and Catalysis by the Histone Demethylase Lsd1-Corest Complex.
Structure, 19, 2011
|
|
6ST3
 
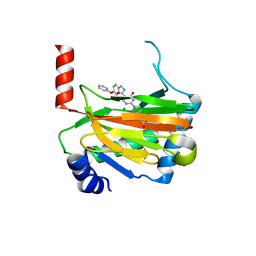 | | HIF prolyl hydroxylase 2 (PHD2/ EGLN1) in complex with 4-hydroxy-N-(4-phenoxybenzyl)-2-(1H-pyrazol-1-yl)pyrimidine-5-carboxamide | | Descriptor: | 4-oxidanyl-~{N}-[(4-phenoxyphenyl)methyl]-2-pyrazol-1-yl-pyrimidine-5-carboxamide, Egl nine homolog 1, FORMIC ACID, ... | | Authors: | Chowdhury, R, Holt-Martyn, J.P, Schofield, C.J. | | Deposit date: | 2019-09-10 | | Release date: | 2019-12-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.426 Å) | | Cite: | Structure-Activity Relationship and Crystallographic Studies on 4-Hydroxypyrimidine HIF Prolyl Hydroxylase Domain Inhibitors.
Chemmedchem, 15, 2020
|
|
3U9Q
 
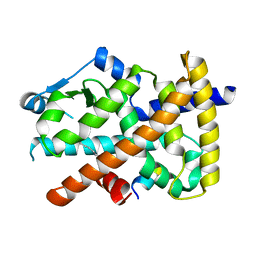 | |
6PPF
 
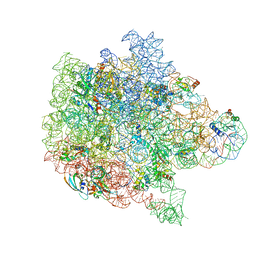 | | Bacterial 45SRbgA ribosomal particle class B | | Descriptor: | 23S rRNA, 50S ribosomal protein L13, 50S ribosomal protein L14, ... | | Authors: | Ortega, J, Seffouh, A, Jain, N, Jahagirdar, D, Basu, K, Razi, A, Ni, X, Guarne, A, Britton, R.A. | | Deposit date: | 2019-07-06 | | Release date: | 2019-09-18 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural consequences of the interaction of RbgA with a 50S ribosomal subunit assembly intermediate.
Nucleic Acids Res., 47, 2019
|
|
6TW1
 
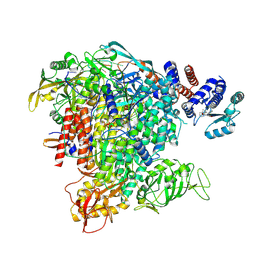 | | Bat Influenza A polymerase termination complex with pyrophosphate using 44-mer vRNA template with mutated oligo(U) sequence | | Descriptor: | 5-oxidanyl-4-oxidanylidene-1-[(1-pyrrolo[2,3-b]pyridin-1-ylcyclopentyl)methyl]pyridine-3-carboxylic acid, MAGNESIUM ION, PYROPHOSPHATE 2-, ... | | Authors: | Wandzik, J.M, Kouba, T, Cusack, S. | | Deposit date: | 2020-01-11 | | Release date: | 2020-04-15 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | A Structure-Based Model for the Complete Transcription Cycle of Influenza Polymerase.
Cell, 2020
|
|
6U2G
 
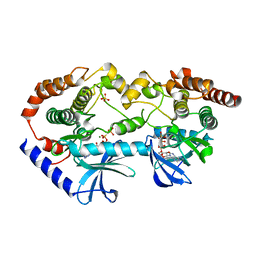 | | BRAF-MEK complex with AMP-PCP bound to BRAF | | Descriptor: | Dual specificity mitogen-activated protein kinase kinase 1, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, ... | | Authors: | Liau, N.P.D, Wendorff, T, Hymowitz, S, Sudhamsu, J. | | Deposit date: | 2019-08-19 | | Release date: | 2019-08-28 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.886 Å) | | Cite: | Negative regulation of RAF kinase activity by ATP is overcome by 14-3-3-induced dimerization.
Nat.Struct.Mol.Biol., 27, 2020
|
|
1LAW
 
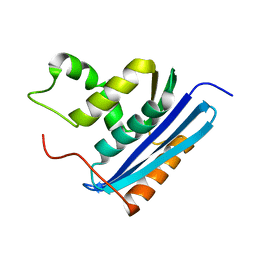 | |
3R2A
 
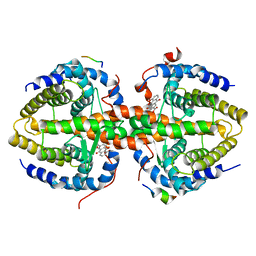 | | Crystal structure of RXRalpha ligand-binding domain complexed with corepressor SMRT2 and antagonist rhein | | Descriptor: | 4,5-dihydroxy-9,10-dioxo-9,10-dihydroanthracene-2-carboxylic acid, Nuclear receptor corepressor 2, Retinoic acid receptor RXR-alpha | | Authors: | Zhang, H, Chen, L, Chen, J, Jiang, H, Shen, X. | | Deposit date: | 2011-03-14 | | Release date: | 2011-05-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis for retinoic x receptor repression on the tetramer.
J.Biol.Chem., 286, 2011
|
|
2XWU
 
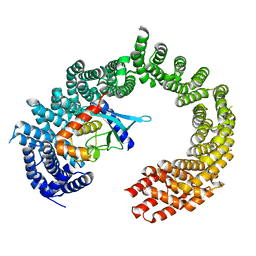 | | CRYSTAL STRUCTURE OF IMPORTIN 13 - UBC9 COMPLEX | | Descriptor: | IMPORTIN13, SUMO-CONJUGATING ENZYME UBC9 | | Authors: | Gruenwald, M, Bono, F. | | Deposit date: | 2010-11-05 | | Release date: | 2010-12-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of Importin13-Ubc9 Complex: Nuclear Import and Release of a Key Regulator of Sumoylation.
Embo J., 30, 2011
|
|
2Y5K
 
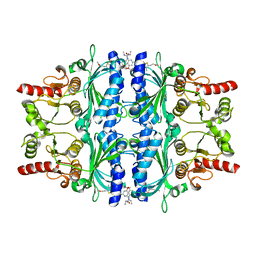 | | Orally active aminopyridines as inhibitors of tetrameric fructose 1,6- bisphosphatase | | Descriptor: | 1-[5-(2-METHOXYETHYL)-4-METHYL-THIOPHEN-2-YL]SULFONYL-3-[4-METHOXY-6-(METHYLCARBAMOYLAMINO)PYRIDIN-2-YL]UREA, FRUCTOSE-1,6-BISPHOSPHATASE 1 | | Authors: | Ruf, A, Hebeisen, P, Haap, W, Kuhn, B, Mohr, P, Wessel, H.P, Zutter, U, Kirchner, S, Benz, J, Joseph, C, Alvarez-Sanchez, R, Gubler, M, Schott, B, Benardeau, A, Tozzo, E, Kitas, E. | | Deposit date: | 2011-01-14 | | Release date: | 2011-05-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Orally Active Aminopyridines as Inhibitors of Tetrameric Fructose-1,6-Bisphosphatase.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
6RVF
 
 | | Crystal structure of hCA II in complex with Urea, N-(1,3-dihydro-1-hydroxy-2,1-benzoxaborol-6-yl)-N'-phenyl | | Descriptor: | 1-[1,1-bis(oxidanyl)-3~{H}-2,1$l^{4}-benzoxaborol-6-yl]-3-phenyl-urea, Carbonic anhydrase 2, ZINC ION | | Authors: | Di Fiore, A, De Simone, G. | | Deposit date: | 2019-05-31 | | Release date: | 2019-08-28 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Exploring benzoxaborole derivatives as carbonic anhydrase inhibitors: a structural and computational analysis reveals their conformational variability as a tool to increase enzyme selectivity.
J Enzyme Inhib Med Chem, 34, 2019
|
|
2V5W
 
 | | Crystal structure of HDAC8-substrate complex | | Descriptor: | GLYCYL-GLYCYL-GLYCINE, HISTONE DEACETYLASE 8, PEPTIDIC SUBSTRATE, ... | | Authors: | Di Marco, S, Vannini, A, Volpari, C. | | Deposit date: | 2007-07-10 | | Release date: | 2007-09-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Substrate Binding to Histone Deacetylases as Revealed by Crystal Structure of Hdac8-Substrate Complex
Embo Rep., 8, 2007
|
|
8ISK
 
 | | Pr conformer of Zea mays phytochrome A1 - ZmphyA1-Pr | | Descriptor: | 3-[5-[[(3~{R},4~{R})-3-ethyl-4-methyl-5-oxidanylidene-3,4-dihydropyrrol-2-yl]methyl]-2-[[5-[(4-ethyl-3-methyl-5-oxidanylidene-pyrrol-2-yl)methyl]-3-(3-hydroxy-3-oxopropyl)-4-methyl-1~{H}-pyrrol-2-yl]methyl]-4-methyl-1~{H}-pyrrol-3-yl]propanoic acid, Phytochrome | | Authors: | Zhang, Y, Ma, C, Zhao, J, Gao, N, Wang, J. | | Deposit date: | 2023-03-20 | | Release date: | 2023-08-09 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural insights into plant phytochrome A as a highly sensitized photoreceptor.
Cell Res., 33, 2023
|
|
8ISJ
 
 | | Pr conformer of Arabidopsis thaliana phytochrome A - AtphyA-Pr | | Descriptor: | 3-[5-[[(3~{R},4~{R})-3-ethyl-4-methyl-5-oxidanylidene-3,4-dihydropyrrol-2-yl]methyl]-2-[[5-[(4-ethyl-3-methyl-5-oxidanylidene-pyrrol-2-yl)methyl]-3-(3-hydroxy-3-oxopropyl)-4-methyl-1~{H}-pyrrol-2-yl]methyl]-4-methyl-1~{H}-pyrrol-3-yl]propanoic acid, Phytochrome A | | Authors: | Zhang, Y, Ma, C, Zhao, J, Gao, N, Wang, J. | | Deposit date: | 2023-03-20 | | Release date: | 2023-08-09 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural insights into plant phytochrome A as a highly sensitized photoreceptor.
Cell Res., 33, 2023
|
|
6V90
 
 | |
