5GJF
 
 | | Crystal structure of human TAK1/TAB1 fusion protein in complex with ligand 3 | | Descriptor: | N-(2-isopropoxy-4-(4-methylpiperazine-1-carbonyl)phenyl)-2-(3-(3-phenylureido)phenyl)thiazole-4-carboxamide, TAK1 kinase - TAB1 chimera fusion protein | | Authors: | Irie, M, Nakamura, M, Fukami, T.A, Matsuura, T, Morishima, K. | | Deposit date: | 2016-06-29 | | Release date: | 2016-11-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Development of a Method for Converting a TAK1 Type I Inhibitor into a Type II or c-Helix-Out Inhibitor by Structure-Based Drug Design (SBDD)
Chem.Pharm.Bull., 64, 2016
|
|
2O9C
 
 | | Crystal Structure of Bacteriophytochrome chromophore binding domain at 1.45 angstrom resolution | | Descriptor: | 3-[2-[(Z)-[3-(2-carboxyethyl)-5-[(Z)-(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-4-methyl-pyrrol-1-ium -2-ylidene]methyl]-5-[(Z)-[(3E)-3-ethylidene-4-methyl-5-oxidanylidene-pyrrolidin-2-ylidene]methyl]-4-methyl-1H-pyrrol-3- yl]propanoic acid, Bacteriophytochrome | | Authors: | Wagner, J.R, Brunzelle, J.S, Vierstra, R.D, Forest, K.T. | | Deposit date: | 2006-12-13 | | Release date: | 2007-03-06 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | High resolution structure of deinococcus bacteriophytochrome yields new insights into phytochrome architecture and evolution.
J.Biol.Chem., 282, 2007
|
|
2OE6
 
 | | 2.4A X-ray crystal structure of unliganded RNA fragment GGGCGUCGCUAGUACC/CGGUACUAAAAGUCGCC containing the human ribosomal decoding A site: RNA construct with 5'-overhang | | Descriptor: | RNA (5'-R(*CP*GP*GP*UP*AP*CP*UP*AP*AP*AP*AP*GP*UP*CP*GP*CP*C)-3'), RNA (5'-R(*GP*GP*GP*CP*GP*UP*CP*GP*CP*UP*AP*GP*UP*AP*CP*C)-3') | | Authors: | Hermann, T, Tereshko, V, Skripkin, E, Patel, D.J. | | Deposit date: | 2006-12-28 | | Release date: | 2007-02-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Apramycin recognition by the human ribosomal decoding site.
Blood Cells Mol.Dis., 38, 2007
|
|
2YIJ
 
 | | Crystal Structure of phospholipase A1 | | Descriptor: | PHOSPHOLIPASE A1-IIGAMMA | | Authors: | Lee, I. | | Deposit date: | 2011-05-13 | | Release date: | 2012-05-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Phospholipase A1
To be Published
|
|
3V76
 
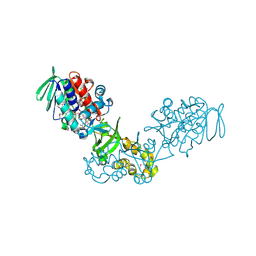 | | The crystal structure of a flavoprotein from Sinorhizobium meliloti | | Descriptor: | DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, Flavoprotein | | Authors: | Zhang, Z, Chamala, S, Evans, B, Foti, R, Gizzi, A, Hillerich, B, Kar, A, LaFleur, J, Seidel, R, Villigas, G, Zencheck, W, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2011-12-20 | | Release date: | 2012-01-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | The crystal structure of a flavoprotein from Sinorhizobium meliloti
TO BE PUBLISHED
|
|
2QYN
 
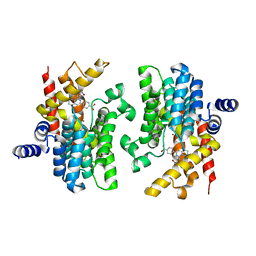 | | Crystal structure of PDE4D2 in complex with inhibitor NPV | | Descriptor: | 4-[8-(3-nitrophenyl)-1,7-naphthyridin-6-yl]benzoic acid, MAGNESIUM ION, ZINC ION, ... | | Authors: | Ke, H. | | Deposit date: | 2007-08-15 | | Release date: | 2008-04-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Structures of the four subfamilies of phosphodiesterase-4 provide insight into the selectivity of their inhibitors.
Biochem.J., 408, 2007
|
|
2ORS
 
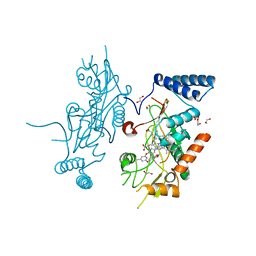 | | Murine Inducible Nitric Oxide Synthase Oxygenase Domain (DELTA 114) 4-(Benzo[1,3]dioxol-5-yloxy)-2-(4-imidazol-1-yl-phenoxy)-6-methyl-pyrimidine Complex | | Descriptor: | 1,2-ETHANEDIOL, 4-(1,3-BENZODIOXOL-5-YLOXY)-2-[4-(1H-IMIDAZOL-1-YL)PHENOXY]-6-METHYLPYRIMIDINE, Nitric oxide synthase, ... | | Authors: | Adler, M, Whitlow, M. | | Deposit date: | 2007-02-04 | | Release date: | 2007-04-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Design, Synthesis, and Activity of 2-Imidazol-1-ylpyrimidine Derived Inducible Nitric Oxide Synthase Dimerization Inhibitors
J.Med.Chem., 50, 2007
|
|
2RS8
 
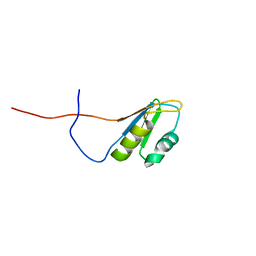 | | Solution structure of the N-terminal RNA recognition motif of NonO | | Descriptor: | Non-POU domain-containing octamer-binding protein | | Authors: | Nagata, T, Muto, Y, Inoue, M, Kigawa, T, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2011-11-29 | | Release date: | 2012-12-19 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the N-terminal RNA recognition motif of NonO
To be Published
|
|
2RCS
 
 | | IMMUNOGLOBULIN 48G7 GERMLINE FAB-AFFINITY MATURATION OF AN ESTEROLYTIC ANTIBODY | | Descriptor: | IMMUNOGLOBULIN 48G7 GERMLINE FAB | | Authors: | Wedemayer, G.J, Wang, L.H, Patten, P.A, Schultz, P.G, Stevens, R.C. | | Deposit date: | 1997-05-14 | | Release date: | 1997-11-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural insights into the evolution of an antibody combining site.
Science, 276, 1997
|
|
3ECN
 
 | | Crystal structure of PDE8A catalytic domain in complex with IBMX | | Descriptor: | 3-ISOBUTYL-1-METHYLXANTHINE, High affinity cAMP-specific and IBMX-insensitive 3',5'-cyclic phosphodiesterase 8A, MAGNESIUM ION, ... | | Authors: | Wang, H, Yan, Z, Yang, S, Cai, J, Robinson, H, Ke, H. | | Deposit date: | 2008-09-01 | | Release date: | 2008-11-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Kinetic and structural studies of phosphodiesterase-8A and implication on the inhibitor selectivity
Biochemistry, 47, 2008
|
|
5ADU
 
 | | The Mechanism of Hydrogen Activation by NiFe-hydrogenases | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, CHLORIDE ION, DODECYL-BETA-D-MALTOSIDE, ... | | Authors: | Evans, R, Brooke, E.J, Wehlin, S.A, Nomerotskaia, E, Sargent, F, Carr, S.B, Phillips, S.E.V, Armstrong, F.A. | | Deposit date: | 2015-08-24 | | Release date: | 2015-11-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Mechanism of hydrogen activation by [NiFe] hydrogenases.
Nat. Chem. Biol., 12, 2016
|
|
2Z23
 
 | | Crystal structure of Y.pestis oligo peptide binding protein OppA with tri-lysine ligand | | Descriptor: | Periplasmic oligopeptide-binding protein, peptide (LYS)(LYS)(LYS) | | Authors: | Tanabe, M, Bertland, T, Mirza, O, Byrne, B, Brown, K.A. | | Deposit date: | 2007-05-17 | | Release date: | 2007-10-30 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of OppA and PstS from Yersinia pestis indicate variability of interactions with transmembrane domains.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
3E8Q
 
 | | X-ray structure of rat arginase I-T135A: the unliganded complex | | Descriptor: | Arginase-1, MANGANESE (II) ION | | Authors: | Shishova, E.Y, Di Costanzo, L, Emig, F.A, Ash, D.E, Christianson, D.W. | | Deposit date: | 2008-08-20 | | Release date: | 2008-12-02 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Probing the specificity determinants of amino acid recognition by arginase.
Biochemistry, 48, 2009
|
|
1EI3
 
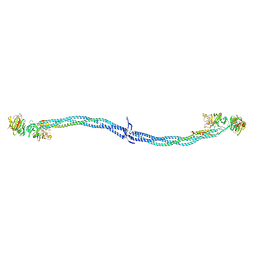 | | CRYSTAL STRUCTURE OF NATIVE CHICKEN FIBRINOGEN | | Descriptor: | FIBRINOGEN | | Authors: | Yang, Z, Mochalkin, I, Veerapandian, L, Riley, M, Doolittle, R.F. | | Deposit date: | 2000-02-23 | | Release date: | 2000-05-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (5.5 Å) | | Cite: | Crystal structure of native chicken fibrinogen at 5.5-A resolution.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
2D0U
 
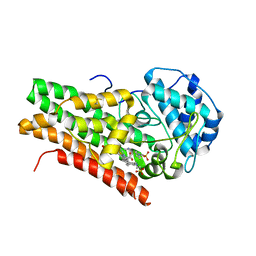 | | Crystal structure of cyanide bound form of human indoleamine 2,3-dioxygenase | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, CYANIDE ION, Indoleamine 2,3-dioxygenase, ... | | Authors: | Sugimoto, H, Oda, S, Otsuki, T, Hino, T, Yoshida, T, Shiro, Y, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-08-08 | | Release date: | 2006-01-31 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Crystal structure of human indoleamine 2,3-dioxygenase: catalytic mechanism of O2 incorporation by a heme-containing dioxygenase.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
1PHZ
 
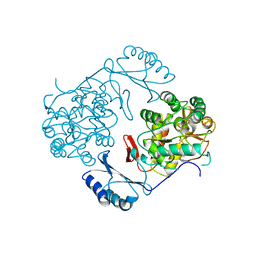 | | STRUCTURE OF PHOSPHORYLATED PHENYLALANINE HYDROXYLASE | | Descriptor: | FE (III) ION, PROTEIN (PHENYLALANINE HYDROXYLASE) | | Authors: | Kobe, B, Jennings, I.G, House, C.M, Michell, B.J, Cotton, R.G, Kemp, B.E. | | Deposit date: | 1998-11-11 | | Release date: | 1999-04-30 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis of autoregulation of phenylalanine hydroxylase.
Nat.Struct.Biol., 6, 1999
|
|
3ECU
 
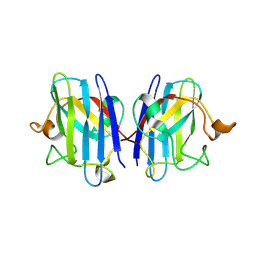 | |
3ECV
 
 | |
3FH7
 
 | |
5SGA
 
 | |
1F2K
 
 | | CRYSTAL STRUCTURE OF ACANTHAMOEBA CASTELLANII PROFILIN II, CUBIC CRYSTAL FORM | | Descriptor: | PROFILIN II | | Authors: | Fedorov, A.A, Shi, W, Mahoney, N, Kaiser, D.A, Almo, S.C. | | Deposit date: | 2000-05-26 | | Release date: | 2000-06-08 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A Comparative Structural Analysis of Profilins
To be Published
|
|
1UE8
 
 | | Crystal Structure of Thermophilic Cytochrome P450 from Sulfolobus tokodaii | | Descriptor: | 367aa long hypothetical cytochrome P450, CHLORIDE ION, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Nakamura, N, Kamitori, S, Ohno, H. | | Deposit date: | 2003-05-09 | | Release date: | 2004-07-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure and direct electrochemistry of cytochrome P450 from the thermoacidophilic crenarchaeon, Sulfolobus tokodaii strain 7
J.Inorg.Biochem., 98, 2004
|
|
5SZ4
 
 | | Carbonic anhydrase IX-mimic in complex with 4-(phenyl)-benzenesulfonamide | | Descriptor: | 4-(phenyl)-benzenesulfonamide, Carbonic anhydrase 2, DIMETHYL SULFOXIDE, ... | | Authors: | Bhatt, A, Mahon, B.P, Cornelio, B, McKenna, R. | | Deposit date: | 2016-08-12 | | Release date: | 2016-12-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure-Activity Relationships of Benzenesulfonamide-Based Inhibitors towards Carbonic Anhydrase Isoform Specificity.
Chembiochem, 18, 2017
|
|
5T4B
 
 | | Human DPP4 in complex with a ligand 34a | | Descriptor: | 2-[(3R)-3-aminopiperidin-1-yl]-3-(but-2-yn-1-yl)-5-[(4-methylquinazolin-2-yl)methyl]-3H-imidazo[2,1-b]purin-4(5H)-one, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Scapin, G. | | Deposit date: | 2016-08-29 | | Release date: | 2016-10-05 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Scaffold-hopping from xanthines to tricyclic guanines: A case study of dipeptidyl peptidase 4 (DPP4) inhibitors.
Bioorg.Med.Chem., 24, 2016
|
|
3FA6
 
 | |
