6M7P
 
 | | Human DNA polymerase eta extension complex with cdA at the -2 position | | Descriptor: | 2'-deoxy-5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]adenosine, DNA (5'-D(*AP*GP*TP*GP*TP*GP*TP*G)-3'), DNA (5'-D(*CP*AP*TP*TP*CP*(02I)P*CP*AP*CP*AP*CP*T)-3'), ... | | Authors: | Gregory, M.T, Yang, W. | | Deposit date: | 2018-08-20 | | Release date: | 2018-09-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Bypassing a 8,5'-cyclo-2'-deoxyadenosine lesion by human DNA polymerase eta at atomic resolution.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6M7V
 
 | | Human DNA polymerase eta extension complex with cdA at the -1 position | | Descriptor: | 2'-deoxy-5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]adenosine, DNA (5'-D(P*(02I)P*CP*TP*CP*AP*CP*AP*CP*T)-3'), DNA (5'-D(P*GP*TP*GP*TP*GP*AP*GP*T)-3'), ... | | Authors: | Weng, P, Gao, Y, Yang, W. | | Deposit date: | 2018-08-21 | | Release date: | 2018-09-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.062 Å) | | Cite: | Bypassing a 8,5'-cyclo-2'-deoxyadenosine lesion by human DNA polymerase eta at atomic resolution.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6M7O
 
 | | Human DNA polymerase eta ternary complex with Mn2+ and dTMPNPP oppositing cdA | | Descriptor: | 5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]thymidine, DNA (5'-D(*AP*TP*(02I)P*CP*TP*CP*AP*CP*AP*CP*T)-3'), DNA (5'-D(P*AP*GP*TP*GP*TP*GP*AP*G*(1FZ))-3'), ... | | Authors: | Weng, P, Gao, Y, Yang, W. | | Deposit date: | 2018-08-20 | | Release date: | 2018-09-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Bypassing a 8,5'-cyclo-2'-deoxyadenosine lesion by human DNA polymerase eta at atomic resolution.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
4PWM
 
 | | Crystal structure of Dickerson Drew Dodecamer with 5-carboxycytosine | | Descriptor: | 5'-[CGCGAATT(5CC)GCG]-3' | | Authors: | Szulik, M.W, Pallan, P, Banerjee, S, Voehler, M, Egli, M, Stone, M.P. | | Deposit date: | 2014-03-20 | | Release date: | 2015-02-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Differential stabilities and sequence-dependent base pair opening dynamics of watson-crick base pairs with 5-hydroxymethylcytosine, 5-formylcytosine, or 5-carboxylcytosine.
Biochemistry, 54, 2015
|
|
6M7T
 
 | | Human DNA polymerase eta in a non-productive ternary complex with Ca2+ and dTTP oppositing cdA | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, DNA (5'-D(*AP*GP*TP*GP*TP*GP*AP*G)-3'), ... | | Authors: | Weng, P, Gao, Y, Yang, W. | | Deposit date: | 2018-08-21 | | Release date: | 2018-09-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Bypassing a 8,5'-cyclo-2'-deoxyadenosine lesion by human DNA polymerase eta at atomic resolution.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
4QC7
 
 | | Dodecamer structure of 5-formylcytosine containing DNA | | Descriptor: | short DNA strands | | Authors: | Szulik, M.W, Pallan, P, Egli, M, Stone, M.P. | | Deposit date: | 2014-05-09 | | Release date: | 2015-02-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Differential stabilities and sequence-dependent base pair opening dynamics of watson-crick base pairs with 5-hydroxymethylcytosine, 5-formylcytosine, or 5-carboxylcytosine.
Biochemistry, 54, 2015
|
|
6M7U
 
 | | Human DNA polymerase eta in a non-productive ternary complex with Mg2+ and dTMPNPP oppositing cdA | | Descriptor: | 5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]thymidine, CHLORIDE ION, DNA (5'-D(*AP*GP*TP*GP*TP*GP*AP*G)-3'), ... | | Authors: | Weng, P, Gao, Y, Yang, W. | | Deposit date: | 2018-08-21 | | Release date: | 2018-09-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Bypassing a 8,5'-cyclo-2'-deoxyadenosine lesion by human DNA polymerase eta at atomic resolution.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
4MWO
 
 | | Crystal structure of human mitochondrial 5'(3')-deoxyribonucleotidase in complex with the inhibitor CPB-T | | Descriptor: | 1-{3,5-O-[(4-carboxyphenyl)(phosphono)methylidene]-2-deoxy-beta-D-threo-pentofuranosyl}-5-methylpyrimidine-2,4(1H,3H)-dione, 5'(3')-deoxyribonucleotidase, mitochondrial, ... | | Authors: | Pachl, P, Rezacova, P, Brynda, J. | | Deposit date: | 2013-09-25 | | Release date: | 2014-09-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Conformationally constrained nucleoside phosphonic acids - potent inhibitors of human mitochondrial and cytosolic 5'(3')-nucleotidases.
Org.Biomol.Chem., 12, 2014
|
|
1DA0
 
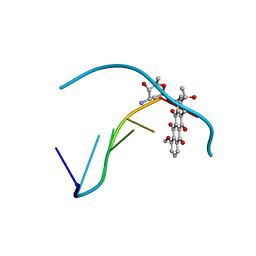 | | DNA-DRUG INTERACTIONS: THE CRYSTAL STRUCTURE OF D(CGATCG) COMPLEXED WITH DAUNOMYCIN | | Descriptor: | DAUNOMYCIN, DNA (5'-D(*CP*GP*AP*TP*CP*G)-3') | | Authors: | Moore, M.H, Hunter, W.N, Langlois D'Estaintot, B, Kennard, O. | | Deposit date: | 1992-10-17 | | Release date: | 1993-07-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | DNA-drug interactions. The crystal structure of d(CGATCG) complexed with daunomycin.
J.Mol.Biol., 206, 1989
|
|
1D90
 
 | | REFINED CRYSTAL STRUCTURE OF AN OCTANUCLEOTIDE DUPLEX WITH I.T MISMATCHED BASE PAIRS | | Descriptor: | DNA (5'-D(*GP*GP*IP*GP*CP*TP*CP*C)-3') | | Authors: | Cruse, W.B.T, Aymani, J, Kennard, O, Brown, T, Jack, A.G.C, Leonard, G.A. | | Deposit date: | 1992-10-17 | | Release date: | 1993-07-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Refined crystal structure of an octanucleotide duplex with I.T. mismatched base pairs.
Nucleic Acids Res., 17, 1989
|
|
2Z70
 
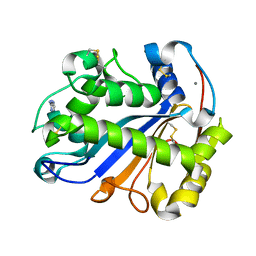 | | E.coli RNase 1 in complex with d(CGCGATCGCG) | | Descriptor: | CALCIUM ION, DNA (5'-D(*DCP*DGP*DCP*DGP*DAP*DTP*DCP*DGP*DCP*DG)-3'), Ribonuclease I | | Authors: | Martinez-Rodriguez, S, Loris, R, Messens, J. | | Deposit date: | 2007-08-09 | | Release date: | 2008-06-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Nonspecific base recognition mediated by water bridges and hydrophobic stacking in ribonuclease I from Escherichia coli
Protein Sci., 17, 2008
|
|
3PF4
 
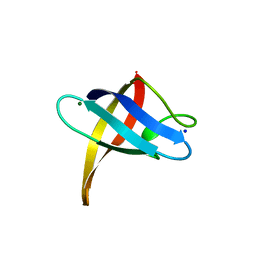 | | Crystal structure of Bs-CspB in complex with r(GUCUUUA) | | Descriptor: | Cold shock protein cspB, MAGNESIUM ION, SODIUM ION, ... | | Authors: | Sachs, R, Max, K.E.A, Heinemann, U. | | Deposit date: | 2010-10-27 | | Release date: | 2011-09-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | RNA single strands bind to a conserved surface of the major cold shock protein in crystals and solution.
Rna, 18, 2012
|
|
3PF5
 
 | |
3RDP
 
 | | Crystal structure of thymidine kinase from herpes simplex virus type 1 in complex with N-METHYL-FHBT | | Descriptor: | 6-[(2R)-2-(fluoromethyl)-3-hydroxy-propyl]-1,5-dimethyl-pyrimidine-2,4-dione, SULFATE ION, Thymidine kinase | | Authors: | Pernot, L, Perozzo, R, Westermaier, Y, Martic, M, Ametamey, S, Scapozza, L. | | Deposit date: | 2011-04-01 | | Release date: | 2011-08-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Synthesis, crystal structure, and in vitro biological evaluation of C-6 pyrimidine derivatives: new lead structures for monitoring gene expression in vivo.
Nucleosides Nucleotides Nucleic Acids, 30, 2011
|
|
3OPI
 
 | | 7-DEAZA-2'-DEOXYADENOSINE modification in B-FORM DNA | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*AP*(7DA)P*TP*TP*CP*GP*CP*G)-3'), MAGNESIUM ION, SODIUM ION | | Authors: | Kowal, E.A, Ganguly, M, Pallan, P.S, Marky, L.A, Gold, B, Egli, M, Stone, M.P. | | Deposit date: | 2010-09-01 | | Release date: | 2011-08-31 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Altering the Electrostatic Potential in the Major Groove: Thermodynamic and Structural Characterization of 7-Deaza-2'-deoxyadenosine:dT Base Pairing in DNA.
J.Phys.Chem.B, 115, 2011
|
|
2DND
 
 | | A BIFURCATED HYDROGEN-BONDED CONFORMATION IN THE D(A.T) BASE PAIRS OF THE DNA DODECAMER D(CGCAAATTTGCG) AND ITS COMPLEX WITH DISTAMYCIN | | Descriptor: | DISTAMYCIN A, DNA (5'-D(*CP*GP*CP*AP*AP*AP*TP*TP*TP*GP*CP*G)-3') | | Authors: | Coll, M, Frederick, C.A, Wang, A.H.-J, Rich, A. | | Deposit date: | 1988-08-29 | | Release date: | 1989-01-09 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A bifurcated hydrogen-bonded conformation in the d(A.T) base pairs of the DNA dodecamer d(CGCAAATTTGCG) and its complex with distamycin.
Proc.Natl.Acad.Sci.USA, 84, 1987
|
|
1MRN
 
 | | CRYSTAL STRUCTURE OF MYCOBACTERIUM TUBERCULOSIS THYMIDYLATE KINASE COMPLEXED WITH BISUBSTRATE INHIBITOR (TP5A) | | Descriptor: | MAGNESIUM ION, P1-(5'-ADENOSYL)P5-(5'-THYMIDYL)PENTAPHOSPHATE, SULFATE ION, ... | | Authors: | Haouz, A, Vanheusden, V, Munier-Lehmann, H, Froeyen, M, Herdewijn, P, Van Calenbergh, S, Delarue, M. | | Deposit date: | 2002-09-18 | | Release date: | 2003-01-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Enzymatic and structural analysis of inhibitors designed against Mycobacterium tuberculosis thymidylate kinase. New insights into the phosphoryl transfer mechanism.
J.Biol.Chem., 278, 2003
|
|
1MRS
 
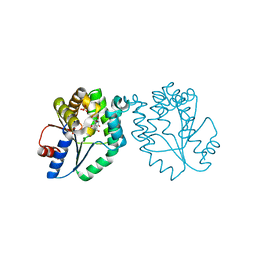 | | CRYSTAL STRUCTURE OF MYCOBACTERIUM TUBERCULOSIS THYMIDYLATE KINASE COMPLEXED WITH 5-CH2OH DEOXYURIDINE MONOPHOSPHATE | | Descriptor: | 5-HYDROXYMETHYLURIDINE-2'-DEOXY-5'-MONOPHOSPHATE, MAGNESIUM ION, SULFATE ION, ... | | Authors: | Haouz, A, Vanheusden, V, Munier-Lehmann, H, Froeyen, M, Herdewijn, P, Van Calenbergh, S, Delarue, M. | | Deposit date: | 2002-09-18 | | Release date: | 2003-01-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Enzymatic and structural analysis of inhibitors designed against Mycobacterium tuberculosis thymidylate kinase. New insights into the phosphoryl transfer mechanism.
J.Biol.Chem., 278, 2003
|
|
4H10
 
 | |
2UVR
 
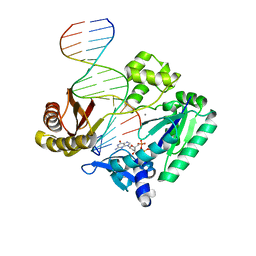 | | Crystal structures of mutant Dpo4 DNA polymerases with 8-oxoG containing DNA template-primer constructs | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, 5'-D(*GP*GP*GP*GP*GP*AP*AP*GP*GP*AP *TP*TP*CP*C)-3', 5'-D(*TP*CP*AP*CP*8OGP*GP*AP*AP*TP*CP *CP*TP*TP*CP*CP*CP*CP*C)-3', ... | | Authors: | Irimia, A, Egli, M. | | Deposit date: | 2007-03-13 | | Release date: | 2007-05-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Hydrogen Bonding of 7,8-Dihydro-8-Oxodeoxyguanosine with a Charged Residue in the Little Finger Domain Determines Miscoding Events in Sulfolobus Solfataricus DNA Polymerase Dpo4.
J.Biol.Chem., 282, 2007
|
|
4HQI
 
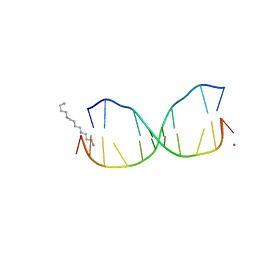 | | Structure of O6-Benzyl-2'-deoxyguanosine opposite perimidinone-derived synthetic nucleoside in DNA duplex | | Descriptor: | SPERMINE, STRONTIUM ION, Short modified nucleic acids | | Authors: | Kowal, E.A, Lad, R, Pallan, P.S, Muffly, E, Wawrzak, Z, Egli, M, Sturla, S.J, Stone, M.P. | | Deposit date: | 2012-10-25 | | Release date: | 2013-07-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Recognition of O6-benzyl-2'-deoxyguanosine by a perimidinone-derived synthetic nucleoside: a DNA interstrand stacking interaction.
Nucleic Acids Res., 41, 2013
|
|
2UVW
 
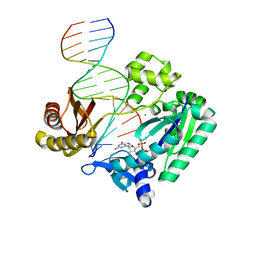 | | Crystal structures of mutant Dpo4 DNA polymerases with 8-oxoG containing DNA template-primer constructs | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, 5'-D(*GP*GP*GP*GP*GP*AP*AP*GP*GP*AP *TP*TP*CP*A)-3', 5'-D(*TP*CP*AP*CP*8OGP*GP*AP*AP*TP*CP *CP*TP*TP*CP*CP*CP*CP*C)-3', ... | | Authors: | Irimia, A, Egli, M. | | Deposit date: | 2007-03-14 | | Release date: | 2007-05-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Hydrogen Bonding of 7,8-Dihydro-8-Oxodeoxyguanosine with a Charged Residue in the Little Finger Domain Determines Miscoding Events in Sulfolobus Solfataricus DNA Polymerase Dpo4.
J.Biol.Chem., 282, 2007
|
|
2UVU
 
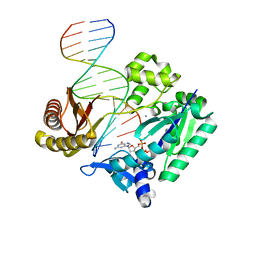 | | Crystal structures of mutant Dpo4 DNA polymerases with 8-oxoG containing DNA template-primer constructs | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, 5'-D(*GP*GP*GP*GP*GP*AP*AP*GP*GP*AP *TP*TP*CP*A)-3', 5'-D(*TP*CP*AP*CP*8OGP*GP*AP*AP*TP*CP *CP*TP*TP*CP*CP*CP*CP*C)-3', ... | | Authors: | Irimia, A, Egli, M. | | Deposit date: | 2007-03-14 | | Release date: | 2007-05-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Hydrogen Bonding of 7,8-Dihydro-8-Oxodeoxyguanosine with a Charged Residue in the Little Finger Domain Determines Miscoding Events in Sulfolobus Solfataricus DNA Polymerase Dpo4.
J.Biol.Chem., 282, 2007
|
|
2UVV
 
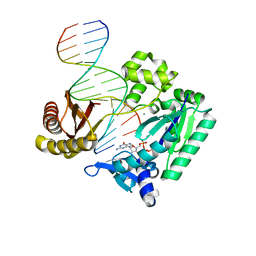 | | Crystal structures of mutant Dpo4 DNA polymerases with 8-oxoG containing DNA template-primer constructs | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, 5'-D(*GP*GP*GP*GP*GP*AP*AP*GP*GP*AP *TP*TP*CP*C)-3', 5'-D(*TP*CP*AP*CP*8OGP*GP*AP*AP*TP*CP *CP*TP*TP*CP*CP*CP*CP*C)-3', ... | | Authors: | Irimia, A, Egli, M. | | Deposit date: | 2007-03-14 | | Release date: | 2007-05-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Hydrogen Bonding of 7,8-Dihydro-8-Oxodeoxyguanosine with a Charged Residue in the Little Finger Domain Determines Miscoding Events in Sulfolobus Solfataricus DNA Polymerase Dpo4.
J.Biol.Chem., 282, 2007
|
|
4ASJ
 
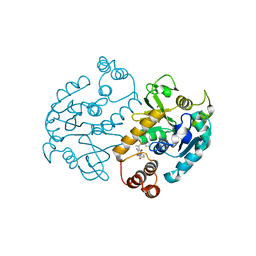 | | Pseudomonas aeruginosa RmlA in complex with allosteric inhibitor | | Descriptor: | CHLORIDE ION, GLUCOSE-1-PHOSPHATE THYMIDYLYLTRANSFERASE, N-(6-amino-1-benzyl-2,4-dioxo-1,2,3,4-tetrahydropyrimidin-5-yl)-N-methylbenzenesulfonamide, ... | | Authors: | Alphey, M.S, Pirrie, L, Torrie, L.S, Gardiner, M, Westwood, N.J, Gray, D, Naismith, J.H. | | Deposit date: | 2012-05-01 | | Release date: | 2012-10-31 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Allosteric competitive inhibitors of the glucose-1-phosphate thymidylyltransferase (RmlA) from Pseudomonas aeruginosa.
ACS Chem. Biol., 8, 2013
|
|
