3RK4
 
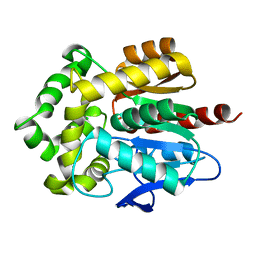 | | Structure of Rhodococcus rhodochrous haloalkane dehalogenase mutant DhaA31 | | Descriptor: | CHLORIDE ION, Haloalkane dehalogenase | | Authors: | Lahoda, M, Stsiapanava, A, Mesters, J, Chaloupkova, R, Damborsky, J, Kuta Smatanova, I. | | Deposit date: | 2011-04-17 | | Release date: | 2012-04-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Crystallographic analysis of 1,2,3-trichloropropane biodegradation by the haloalkane dehalogenase DhaA31.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
5RW2
 
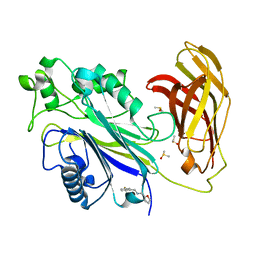 | | INPP5D PanDDA analysis group deposition -- Crystal Structure of the phosphatase and C2 domains of SHIP1 in complex with Z2737076969 | | Descriptor: | 1-(3-fluorophenyl)-N-[(furan-2-yl)methyl]methanamine, DIMETHYL SULFOXIDE, Phosphatidylinositol 3,4,5-trisphosphate 5-phosphatase 1 | | Authors: | Bradshaw, W.J, Newman, J.A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Gileadi, O. | | Deposit date: | 2020-10-30 | | Release date: | 2020-11-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | Regulation of inositol 5-phosphatase activity by the C2 domain of SHIP1 and SHIP2.
Structure, 2024
|
|
1JAE
 
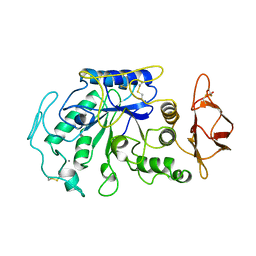 | | STRUCTURE OF TENEBRIO MOLITOR LARVAL ALPHA-AMYLASE | | Descriptor: | ALPHA-AMYLASE, CALCIUM ION, CHLORIDE ION | | Authors: | Strobl, S, Maskos, K, Betz, M, Wiegand, G, Huber, R, Gomis-Rueth, F.X, Frank, G, Glockshuber, R. | | Deposit date: | 1997-09-30 | | Release date: | 1998-11-04 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of yellow meal worm alpha-amylase at 1.64 A resolution.
J.Mol.Biol., 278, 1998
|
|
5RXC
 
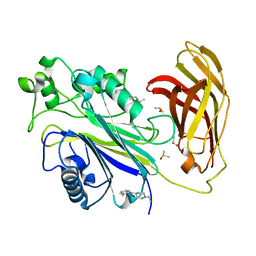 | | INPP5D PanDDA analysis group deposition -- Crystal Structure of the phosphatase and C2 domains of SHIP1 in complex with Z2856434868 | | Descriptor: | 1-ethyl-N-(2-fluorophenyl)piperidin-4-amine, DIMETHYL SULFOXIDE, Phosphatidylinositol 3,4,5-trisphosphate 5-phosphatase 1 | | Authors: | Bradshaw, W.J, Newman, J.A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Gileadi, O. | | Deposit date: | 2020-10-30 | | Release date: | 2020-11-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Regulation of inositol 5-phosphatase activity by the C2 domain of SHIP1 and SHIP2.
Structure, 2024
|
|
1CKP
 
 | | HUMAN CYCLIN DEPENDENT KINASE 2 COMPLEXED WITH THE INHIBITOR PURVALANOL B | | Descriptor: | 1,2-ETHANEDIOL, PROTEIN (CYCLIN-DEPENDENT PROTEIN KINASE 2), PURVALANOL B | | Authors: | Gray, N.S, Thunnissen, A.M.W.H, Schultz, P.G, Kim, S.H. | | Deposit date: | 1998-07-14 | | Release date: | 1999-01-13 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Exploiting chemical libraries, structure, and genomics in the search for kinase inhibitors.
Science, 281, 1998
|
|
5RFT
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 main protease in complex with PCM-0102432 | | Descriptor: | 1-[(4S)-4-phenyl-3,4-dihydroisoquinolin-2(1H)-yl]ethan-1-one, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Owen, C.D, Douangamath, A, Lukacik, P, Powell, A.J, Strain-Damerell, C.M, Resnick, E, Krojer, T, Gehrtz, P, Wild, C, Aimon, A, Brandao-Neto, J, Carbery, A, Dunnett, L, Skyner, R, Snee, M, London, N, Walsh, M.A, von Delft, F. | | Deposit date: | 2020-03-15 | | Release date: | 2020-03-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Crystallographic and electrophilic fragment screening of the SARS-CoV-2 main protease.
Nat Commun, 11, 2020
|
|
2YSD
 
 | | Solution structure of the first WW domain from the human membrane-associated guanylate kinase, WW and PDZ domain-containing protein 1. MAGI-1 | | Descriptor: | Membrane-associated guanylate kinase, WW and PDZ domain-containing protein 1 | | Authors: | Ohnishi, S, Tochio, N, Koshiba, S, Harada, T, Watanabe, S, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-03 | | Release date: | 2007-10-09 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the first WW domain from the human membrane-associated guanylate kinase, WW and PDZ domain-containing protein 1. MAGI-1
To be Published
|
|
3GY4
 
 | | A comparative study on the inhibition of bovine beta-trypsin by bis-benzamidines diminazene and pentamidine by X-ray crystallography and ITC | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Cationic trypsin, ... | | Authors: | Perilo, C.S, Pereira, M.T, Santoro, M.M, Nagem, R.A.P. | | Deposit date: | 2009-04-03 | | Release date: | 2010-03-23 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural binding evidence of the trypanocidal drugs Berenil and Pentacarinate active principles to a serine protease model.
Int.J.Biol.Macromol., 46, 2010
|
|
3R2N
 
 | |
2TRX
 
 | | CRYSTAL STRUCTURE OF THIOREDOXIN FROM ESCHERICHIA COLI AT 1.68 ANGSTROMS RESOLUTION | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, COPPER (II) ION, THIOREDOXIN | | Authors: | Katti, S.K, Lemaster, D.M, Eklund, H. | | Deposit date: | 1990-03-19 | | Release date: | 1991-10-15 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Crystal structure of thioredoxin from Escherichia coli at 1.68 A resolution.
J.Mol.Biol., 212, 1990
|
|
3RRQ
 
 | |
3R9T
 
 | |
4F74
 
 | |
3RHK
 
 | | Crystal structure of the catalytic domain of c-Met kinase in complex with ARQ 197 | | Descriptor: | 1-[(3R,4R)-4-(1H-indol-3-yl)-2,5-dioxopyrrolidin-3-yl]pyrrolo[3,2,1-ij]quinolinium, Hepatocyte growth factor receptor | | Authors: | Eathiraj, S, Palma, R, Volckova, E, Hirschi, M, France, D.S, Ashwell, M.A, Chan, T.C. | | Deposit date: | 2011-04-11 | | Release date: | 2011-04-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Discovery of a novel mode of protein kinase inhibition characterized by the mechanism of inhibition of human mesenchymal-epithelial transition factor (c-Met) protein autophosphorylation by ARQ 197.
J.Biol.Chem., 286, 2011
|
|
4A2Z
 
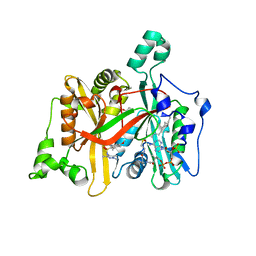 | | CRYSTAL STRUCTURE OF LEISHMANIA MAJOR N-MYRISTOYLTRANSFERASE (NMT) WITH BOUND MYRISTOYL-COA AND A PYRAZOLE SULPHONAMIDE LIGAND | | Descriptor: | 4-METHOXY-2,3,6-TRIMETHYL-N-(1,3,5-TRIMETHYL-1H-PYRAZOL-4-YL)BENZENESULFONAMIDE, CHLORIDE ION, GLYCEROL, ... | | Authors: | Robinson, D.A, Brand, S, Fairlamb, A.H, Ferguson, M.A.J, Frearson, J.A, Wyatt, P.G. | | Deposit date: | 2011-09-29 | | Release date: | 2011-12-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Discovery of a novel class of orally active trypanocidal N-myristoyltransferase inhibitors.
J. Med. Chem., 55, 2012
|
|
4AMC
 
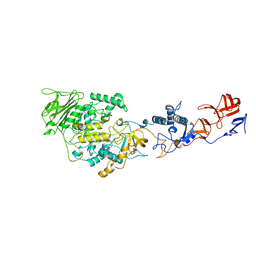 | | Crystal structure of Lactobacillus reuteri 121 N-terminally truncated glucansucrase GTFA | | Descriptor: | CALCIUM ION, GLUCANSUCRASE | | Authors: | Pijning, T, Vujicic-Zagar, A, Kralj, S, Dijkhuizen, L, Dijkstra, B.W. | | Deposit date: | 2012-03-08 | | Release date: | 2012-11-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structure of the Alpha-1,6/Alpha-1,4-Specific Glucansucrase Gtfa from Lactobacillus Reuteri 121
Acta Crystallogr.,Sect.F, 68, 2012
|
|
3CJO
 
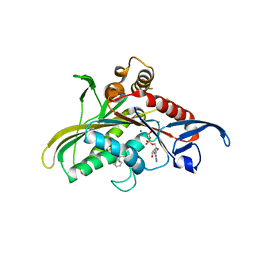 | | Crystal structure of KSP in complex with inhibitor 30 | | Descriptor: | (2S)-4-(2,5-difluorophenyl)-N-[(3R,4S)-3-fluoro-1-methylpiperidin-4-yl]-2-(hydroxymethyl)-N-methyl-2-phenyl-2,5-dihydro-1H-pyrrole-1-carboxamide, ADENOSINE-5'-DIPHOSPHATE, Kinesin-like protein KIF11, ... | | Authors: | Yan, Y. | | Deposit date: | 2008-03-13 | | Release date: | 2008-07-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Kinesin spindle protein (KSP) inhibitors. 9. Discovery of (2S)-4-(2,5-difluorophenyl)-n-[(3R,4S)-3-fluoro-1-methylpiperidin-4-yl]-2-(hydroxymethyl)-N-methyl-2-phenyl-2,5-dihydro-1H-pyrrole-1-carboxamide (MK-0731) for the treatment of taxane-refractory cancer.
J.Med.Chem., 51, 2008
|
|
2C31
 
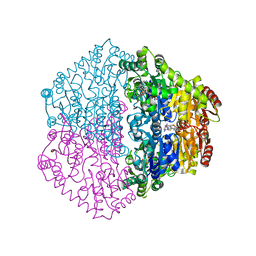 | | CRYSTAL STRUCTURE OF OXALYL-COA DECARBOXYLASE IN COMPLEX WITH THE COFACTOR DERIVATIVE THIAMIN-2-THIAZOLONE DIPHOSPHATE AND ADENOSINE DIPHOSPHATE | | Descriptor: | 2-{3-[(4-AMINO-2-METHYLPYRIMIDIN-5-YL)METHYL]-4-METHYL-2-OXO-2,3-DIHYDRO-1,3-THIAZOL-5-YL}ETHYL TRIHYDROGEN DIPHOSPHATE, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Berthold, C.L, Moussatche, P, Richards, N.G.J, Lindqvist, Y. | | Deposit date: | 2005-10-03 | | Release date: | 2005-10-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Structural Basis for Activation of the Thiamin Diphosphate-Dependent Enzyme Oxalyl-Coa Decarboxylase by Adenosine Diphosphate.
J.Biol.Chem., 280, 2005
|
|
7KP6
 
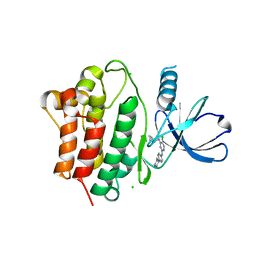 | | Structure of Ack1 kinase in complex with a selective inhibitor | | Descriptor: | 5-chloro-N~2~-[4-(4-methylpiperazin-1-yl)phenyl]-N~4~-{[(2R)-oxolan-2-yl]methyl}pyrimidine-2,4-diamine, Activated CDC42 kinase 1, CHLORIDE ION | | Authors: | Thakur, M.K, Miller, W.T, Mahajan, N, Seeliger, M.A. | | Deposit date: | 2020-11-10 | | Release date: | 2022-02-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Inhibiting ACK1-mediated phosphorylation of C-terminal Src kinase counteracts prostate cancer immune checkpoint blockade resistance.
Nat Commun, 13, 2022
|
|
3D39
 
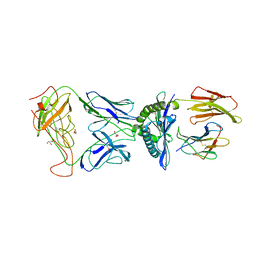 | | The complex between TCR A6 and human Class I MHC HLA-A2 with the modified HTLV-1 TAX (Y5(4-fluoroPhenylalanine)) peptide | | Descriptor: | A6 TCR alpha chain, A6 TCR beta chain, Beta-2-microglobulin, ... | | Authors: | Borbulevych, O.Y, Clemens, J.R, Baker, B.M. | | Deposit date: | 2008-05-09 | | Release date: | 2009-06-16 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Fluorine substitutions in an antigenic peptide selectively modulate T-cell receptor binding in a minimally perturbing manner
Biochem.J., 423, 2009
|
|
5T5L
 
 | | LECTIN FROM BAUHINIA FORFICATA IN COMPLEX WITH TN-PEPTIDE | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-alpha-D-galactopyranose, CALCIUM ION, ... | | Authors: | Lubkowski, J, Wlodawer, A. | | Deposit date: | 2016-08-31 | | Release date: | 2016-12-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.17 Å) | | Cite: | Structural analysis and unique molecular recognition properties of a Bauhinia forficata lectin that inhibits cancer cell growth.
FEBS J., 284, 2017
|
|
5TE4
 
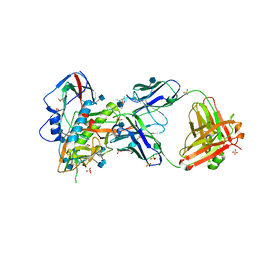 | | Crystal Structure of Broadly Neutralizing VRC01-class Antibody N6 in Complex with HIV-1 Clade G Strain X2088 gp120 Core | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, (R,R)-2,3-BUTANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhou, T, Kwong, P.D. | | Deposit date: | 2016-09-20 | | Release date: | 2016-11-23 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Identification of a CD4-Binding-Site Antibody to HIV that Evolved Near-Pan Neutralization Breadth.
Immunity, 45, 2016
|
|
3CK1
 
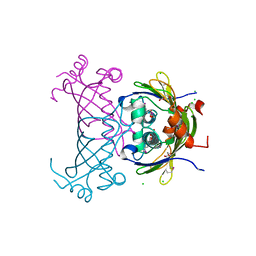 | |
2OCZ
 
 | | The Structure of a Putative 3-Dehydroquinate Dehydratase from Streptococcus pyogenes. | | Descriptor: | 1,2-ETHANEDIOL, 3-dehydroquinate dehydratase, MAGNESIUM ION | | Authors: | Cuff, M.E, Duggan, E, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-12-21 | | Release date: | 2007-01-23 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The Structure of a Putative 3-Dehydroquinate Dehydratase from Streptococcus pyogenes.
TO BE PUBLISHED
|
|
1P7K
 
 | | Crystal structure of an anti-ssDNA antigen-binding fragment (Fab) bound to 4-(2-Hydroxyethyl)piperazine-1-ethanesulfonic acid (HEPES) | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Schuermann, J.P, Henzl, M.T, Deutscher, S.L, Tanner, J.J. | | Deposit date: | 2003-05-02 | | Release date: | 2004-05-11 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure of an anti-DNA fab complexed with a non-DNA ligand provides insights into cross-reactivity and molecular mimicry.
Proteins, 57, 2004
|
|
