7Q72
 
 | | Structure of Pla1 in complex with Red1 | | Descriptor: | NURS complex subunit red1, Poly(A) polymerase pla1 | | Authors: | Soni, K, Wild, K, Sinning, I. | | Deposit date: | 2021-11-09 | | Release date: | 2022-12-07 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Mechanistic insights into RNA surveillance by the canonical poly(A) polymerase Pla1 of the MTREC complex.
Nat Commun, 14, 2023
|
|
7Q73
 
 | | Structure of Pla1 apo | | Descriptor: | GLYCEROL, Poly(A) polymerase pla1 | | Authors: | Soni, K, Wild, K, Sinning, I. | | Deposit date: | 2021-11-09 | | Release date: | 2022-12-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Mechanistic insights into RNA surveillance by the canonical poly(A) polymerase Pla1 of the MTREC complex.
Nat Commun, 14, 2023
|
|
7Q74
 
 | |
8G99
 
 | | Partial auto-inhibitory complex of Xenopus laevis DNA polymerase alpha-primase | | Descriptor: | DNA polymerase alpha catalytic subunit, DNA polymerase alpha subunit B, DNA primase large subunit, ... | | Authors: | Mullins, E.A, Chazin, W.J, Eichman, B.F. | | Deposit date: | 2023-02-21 | | Release date: | 2023-04-12 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | A mechanistic model of primer synthesis from catalytic structures of DNA polymerase alpha-primase.
Nat.Struct.Mol.Biol., 31, 2024
|
|
7OMG
 
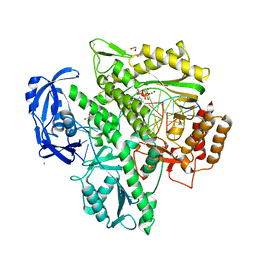 | | Crystal structure of KOD DNA Polymerase in a ternary complex with an Uracil containing template | | Descriptor: | 1,2-ETHANEDIOL, 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, CALCIUM ION, ... | | Authors: | Betz, K, Kropp, H.M, Diederichs, K, Marx, A. | | Deposit date: | 2021-05-22 | | Release date: | 2021-10-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Basis for The Recognition of Deaminated Nucleobases by An Archaeal DNA Polymerase.
Chembiochem, 22, 2021
|
|
7OMB
 
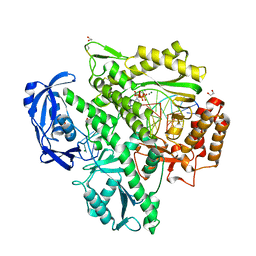 | | Crystal structure of KOD DNA Polymerase in a ternary complex with a p/t duplex containing an extended 5' single stranded template overhang | | Descriptor: | 1,2-ETHANEDIOL, 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, DNA polymerase, ... | | Authors: | Betz, K, Kropp, H.M, Diederichs, K, Marx, A. | | Deposit date: | 2021-05-21 | | Release date: | 2021-10-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structural Basis for The Recognition of Deaminated Nucleobases by An Archaeal DNA Polymerase.
Chembiochem, 22, 2021
|
|
3O2Q
 
 | |
1L3S
 
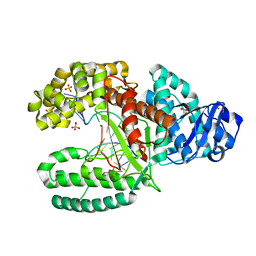 | | Crystal Structure of Bacillus DNA Polymerase I Fragment complexed to 9 base pairs of duplex DNA. | | Descriptor: | 5'-D(*GP*A*CP*GP*TP*AP*CP*GP*TP*GP*AP*TP*CP*GP*CP*A)-3', 5'-D(*GP*CP*GP*AP*TP*CP*AP*CP*G)-3', DNA Polymerase I, ... | | Authors: | Johnson, S.J, Taylor, J.S, Beese, L.S. | | Deposit date: | 2002-03-01 | | Release date: | 2003-03-25 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Processive DNA synthesis observed in a polymerase crystal suggests a
mechanism for the prevention of frameshift mutations
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1L3T
 
 | | Crystal Structure of Bacillus DNA Polymerase I Fragment product complex with 10 base pairs of duplex DNA following addition of a single dTTP residue | | Descriptor: | 5'-D(*GP*AP*CP*G*TP*AP*CP*GP*TP*GP*AP*TP*CP*GP*CP*A)-3', 5'-D(*GP*CP*GP*AP*TP*CP*AP*CP*GP*T)-3', DNA Polymerase I, ... | | Authors: | Johnson, S.J, Taylor, J.S, Beese, L.S. | | Deposit date: | 2002-03-01 | | Release date: | 2003-03-25 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Processive DNA synthesis observed in a polymerase crystal suggests a
mechanism for the prevention of frameshift mutations
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1L3V
 
 | | Crystal Structure of Bacillus DNA Polymerase I Fragment product complex with 15 base pairs of duplex DNA following addition of dTTP, dATP, dCTP, and dGTP residues. | | Descriptor: | 5'-D(*GP*AP*CP*GP*TP*AP*CP*GP*TP*GP*AP*TP*CP*GP*CP*A)-3', 5'-D(*GP*CP*GP*AP*TP*CP*AP*CP*GP*TP*AP*CP*GP*TP*C)-3', DNA Polymerase I, ... | | Authors: | Johnson, S.J, Taylor, J.S, Beese, L.S. | | Deposit date: | 2002-03-01 | | Release date: | 2003-03-25 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Processive DNA synthesis observed in a polymerase crystal suggests a
mechanism for the prevention of frameshift mutations
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
7N4E
 
 | | Escherichia coli sigma 70-dependent paused transcription elongation complex | | Descriptor: | 5'-R(*UP*UP*CP*GP*GP*AP*GP*AP*GP*GP*UP*A)-3', DNA (61-MER), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Molodtsov, V, Su, M, Ebright, R.H. | | Deposit date: | 2021-06-04 | | Release date: | 2022-06-15 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural and mechanistic basis of sigma-dependent transcriptional pausing.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
5KI6
 
 | |
1L3U
 
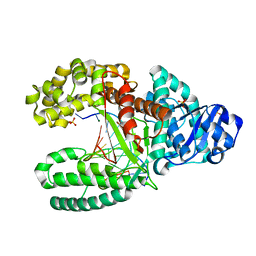 | | Crystal Structure of Bacillus DNA Polymerase I Fragment product complex with 11 base pairs of duplex DNA following addition of a dTTP and a dATP residue. | | Descriptor: | 5'-D(*GP*AP*C*GP*TP*AP*CP*GP*TP*GP*AP*TP*CP*GP*CP*A)-3', 5'-D(*GP*CP*GP*AP*TP*CP*AP*CP*GP*TP*A)-3', DNA Polymerase I, ... | | Authors: | Johnson, S.J, Taylor, J.S, Beese, L.S. | | Deposit date: | 2002-03-01 | | Release date: | 2003-03-25 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Processive DNA synthesis observed in a polymerase crystal suggests a
mechanism for the prevention of frameshift mutations
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1L5U
 
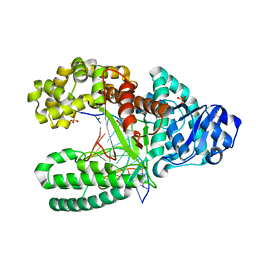 | | Crystal Structure of Bacillus DNA Polymerase I Fragment product complex with 12 base pairs of duplex DNA following addition of a dTTP, a dATP, and a dCTP residue. | | Descriptor: | 5'-D(*GP*A*CP*GP*TP*AP*CP*GP*TP*GP*AP*TP*CP*GP*CP*A)-3', 5'-D(*GP*CP*GP*AP*TP*CP*AP*CP*GP*TP*AP*C)-3', DNA Polymerase I, ... | | Authors: | Johnson, S.J, Taylor, J.S, Beese, L.S. | | Deposit date: | 2002-03-08 | | Release date: | 2003-03-25 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Processive DNA synthesis observed in a polymerase crystal suggests a
mechanism for the prevention of frameshift mutations
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
5C2Y
 
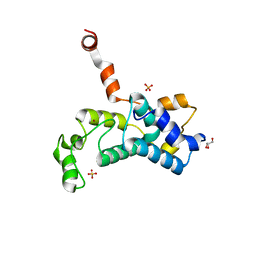 | | Crystal structure of the Saccharomyces cerevisiae Rtr1 (regulator of transcription) | | Descriptor: | GLYCEROL, RNA polymerase II subunit B1 CTD phosphatase RTR1, SULFATE ION, ... | | Authors: | Yogesha, S.D, Irani, S, Zhang, Y.J. | | Deposit date: | 2015-06-16 | | Release date: | 2016-05-04 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of Saccharomyces cerevisiae Rtr1 reveals an active site for an atypical phosphatase.
Sci.Signal., 9, 2016
|
|
8ASD
 
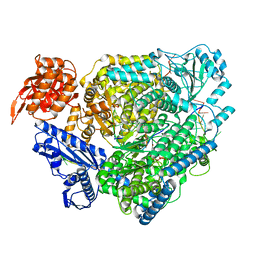 | | Structure of the SFTSV L protein stalled at late elongation [LATE-ELONGATION] | | Descriptor: | 5'-O-[(S)-hydroxy{[(S)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]uridine, MAGNESIUM ION, RNA (5'-R(P*AP*AP*AP*AP*AP*AP*GP*AP*UP*CP*UP*GP*GP*GP*CP*GP*GP*UP*CP*UP*UP*UP*GP*UP*GP*U)-3'), ... | | Authors: | Williams, H.M, Thorkelsson, S.R, Vogel, D, Milewski, M, Busch, C, Cusack, S, Grunewald, K, Quemin, E.R.J, Rosenthal, M. | | Deposit date: | 2022-08-19 | | Release date: | 2023-01-18 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structural insights into viral genome replication by the severe fever with thrombocytopenia syndrome virus L protein.
Nucleic Acids Res., 51, 2023
|
|
1MUK
 
 | | reovirus lambda3 native structure | | Descriptor: | MINOR CORE PROTEIN LAMBDA 3 | | Authors: | Tao, Y, Farsetta, D.L, Nibert, M.L, Harrison, S.C. | | Deposit date: | 2002-09-24 | | Release date: | 2002-12-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | RNA Synthesis in a Cage-Structural Studies of Reovirus Polymerase lambda3
Cell(Cambridge,Mass.), 111, 2002
|
|
2XC9
 
 | | BINARY COMPLEX OF SULFOLOBUS SOLFATARICUS DPO4 DNA POLYMERASE AND 1, N2-ETHENOGUANINE MODIFIED DNA, MAGNESIUM FORM | | Descriptor: | 5'-D(*GP*GP*GP*GP*GP*AP*AP*GP*GP*AP*TP*TP*CP*G)-3', 5'-D(*TP*CP*AP*CP*GNEP*GP*AP*AP*TP*CP*CP*TP*TP* CP*CP*CP*CP*C)-3', DNA POLYMERASE IV | | Authors: | Irimia, A, Loukachevitch, L.V, Egli, M. | | Deposit date: | 2010-04-20 | | Release date: | 2010-09-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Metal Ion Dependence of the Active Site Conformation of the Trans-Lesion DNA Polymerase Dpo4 from Sulfolobus Solfataricus
Acta Crystallogr.,Sect.F, 66, 2010
|
|
5ZAK
 
 | | Cryo-EM structure of human Dicer and its complexes with a pre-miRNA substrate | | Descriptor: | Endoribonuclease Dicer, RISC-loading complex subunit TARBP2 | | Authors: | Liu, Z, Wang, J, Cheng, H, Ke, X, Sun, L, Zhang, Q.C, Wang, H.-W. | | Deposit date: | 2018-02-07 | | Release date: | 2018-05-09 | | Last modified: | 2019-11-06 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Cryo-EM Structure of Human Dicer and Its Complexes with a Pre-miRNA Substrate.
Cell, 173, 2018
|
|
7KPX
 
 | | Structure of the yeast CKM | | Descriptor: | Mediator of RNA polymerase II transcription subunit 12, Mediator of RNA polymerase II transcription subunit 13, Meiotic mRNA stability protein kinase SSN3, ... | | Authors: | Li, Y.C, Chao, T.C, Kim, H.J, Cholko, T, Chen, S.F, Nakanishi, K, Chang, C.E, Murakami, K, Garcia, B.A, Boyer, T.G, Tsai, K.L. | | Deposit date: | 2020-11-12 | | Release date: | 2021-01-27 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Structure and noncanonical Cdk8 activation mechanism within an Argonaute-containing Mediator kinase module.
Sci Adv, 7, 2021
|
|
7KPV
 
 | | Structure of kinase and Central lobes of yeast CKM | | Descriptor: | Mediator of RNA polymerase II transcription subunit 12, Mediator of RNA polymerase II transcription subunit 13, Meiotic mRNA stability protein kinase SSN3, ... | | Authors: | Li, Y.C, Chao, T.C, Kim, H.J, Cholko, T, Chen, S.F, Nakanishi, K, Chang, C.E, Murakami, K, Garcia, B.A, Boyer, T.G, Tsai, K.L. | | Deposit date: | 2020-11-12 | | Release date: | 2021-01-27 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure and noncanonical Cdk8 activation mechanism within an Argonaute-containing Mediator kinase module.
Sci Adv, 7, 2021
|
|
4DRW
 
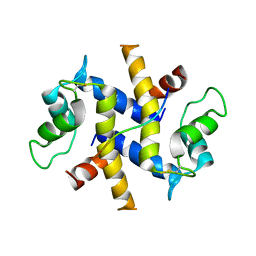 | | Crystal Structure of the Ternary Complex between S100A10, an Annexin A2 N-terminal Peptide and an AHNAK Peptide | | Descriptor: | Neuroblast differentiation-associated protein AHNAK, Protein S100-A10/Annexin A2 chimeric protein | | Authors: | Rezvanpour, A, Lee, T.-W, Junop, M.S, Shaw, G.S. | | Deposit date: | 2012-02-17 | | Release date: | 2012-10-24 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structure of an asymmetric ternary protein complex provides insight for membrane interaction.
Structure, 20, 2012
|
|
7UO4
 
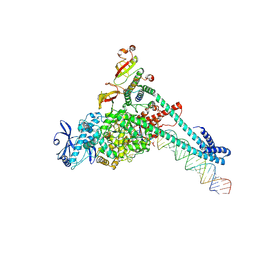 | | SARS-CoV-2 replication-transcription complex bound to Remdesivir triphosphate, in a pre-catalytic state | | Descriptor: | MAGNESIUM ION, Non-structural protein 7, Non-structural protein 8, ... | | Authors: | Malone, B.F, Perry, J.K, Appleby, T.C, Feng, J.Y, Campbell, E.A, Darst, S.A. | | Deposit date: | 2022-04-12 | | Release date: | 2022-11-30 | | Last modified: | 2023-03-08 | | Method: | ELECTRON MICROSCOPY (3.38 Å) | | Cite: | Structural basis for substrate selection by the SARS-CoV-2 replicase.
Nature, 614, 2023
|
|
7UO9
 
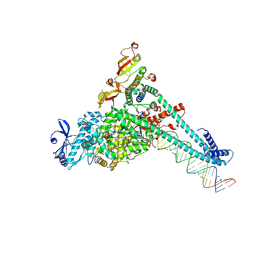 | | SARS-CoV-2 replication-transcription complex bound to UTP, in a pre-catalytic state | | Descriptor: | MAGNESIUM ION, Non-structural protein 7, Non-structural protein 8, ... | | Authors: | Malone, B.F, Perry, J.K, Appleby, T.C, Feng, J.Y, Campbell, E.A, Darst, S.A. | | Deposit date: | 2022-04-12 | | Release date: | 2022-11-30 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.13 Å) | | Cite: | Structural basis for substrate selection by the SARS-CoV-2 replicase.
Nature, 614, 2023
|
|
7UOB
 
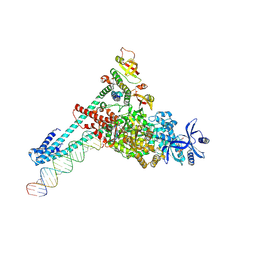 | | SARS-CoV-2 replication-transcription complex bound to GTP, in a pre-catalytic state | | Descriptor: | 3'-DEOXYURIDINE-5'-MONOPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Malone, B.F, Perry, J.K, Appleby, T.C, Feng, J.Y, Campbell, E.A, Darst, S.A. | | Deposit date: | 2022-04-12 | | Release date: | 2022-11-30 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.68 Å) | | Cite: | Structural basis for substrate selection by the SARS-CoV-2 replicase.
Nature, 614, 2023
|
|
