1BUW
 
 | |
1BYA
 
 | | CRYSTAL STRUCTURES OF SOYBEAN BETA-AMYLASE REACTED WITH BETA-MALTOSE AND MALTAL: ACTIVE SITE COMPONENTS AND THEIR APPARENT ROLE IN CATALYSIS | | Descriptor: | BETA-AMYLASE, SULFATE ION | | Authors: | Mikami, B, Degano, M, Hehre, E.J, Sacchettini, J.C. | | Deposit date: | 1994-01-25 | | Release date: | 1994-07-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of soybean beta-amylase reacted with beta-maltose and maltal: active site components and their apparent roles in catalysis.
Biochemistry, 33, 1994
|
|
1BXY
 
 | | CRYSTAL STRUCTURE OF RIBOSOMAL PROTEIN L30 FROM THERMUS THERMOPHILUS AT 1.9 A RESOLUTION: CONFORMATIONAL FLEXIBILITY OF THE MOLECULE. | | Descriptor: | PROTEIN (RIBOSOMAL PROTEIN L30) | | Authors: | Fedorov, R, Nevskaya, N, Khairullina, A, Tishchenko, S, Mikhailov, A, Garber, M, Nikonov, S. | | Deposit date: | 1998-10-09 | | Release date: | 1998-10-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of ribosomal protein L30 from Thermus thermophilus at 1.9 A resolution: conformational flexibility of the molecule.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
1BYR
 
 | |
5MP9
 
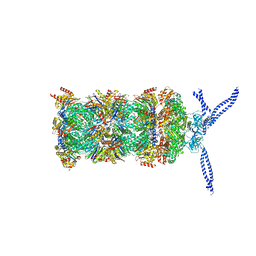 | | 26S proteasome in presence of ATP (s1) | | Descriptor: | 26S protease regulatory subunit 4 homolog, 26S protease regulatory subunit 6A, 26S protease regulatory subunit 6B homolog, ... | | Authors: | Wehmer, M, Rudack, T, Beck, F, Aufderheide, A, Pfeifer, G, Plitzko, J.M, Foerster, F, Schulten, K, Baumeister, W, Sakata, E. | | Deposit date: | 2016-12-16 | | Release date: | 2017-03-08 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structural insights into the functional cycle of the ATPase module of the 26S proteasome.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
1BY2
 
 | |
1BZ7
 
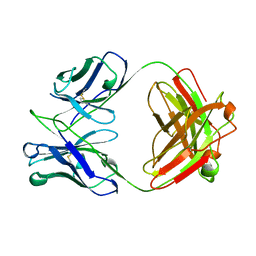 | | FAB FRAGMENT FROM MURINE ASCITES | | Descriptor: | PROTEIN (ANTIBODY R24 (HEAVY CHAIN)), PROTEIN (ANTIBODY R24 (LIGHT CHAIN)) | | Authors: | Kaminski, M.J, Mackenzie, C.R, Mooibroek, M.J, Dahms, T.E.S, Hirama, T, Houghton, A.N, Chapman, P.B, Evans, S.V. | | Deposit date: | 1998-11-06 | | Release date: | 1999-11-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The role of homophilic binding in anti-tumor antibody R24 recognition of molecular surfaces. Demonstration of an intermolecular beta-sheet interaction between vh domains.
J.Biol.Chem., 274, 1999
|
|
1BY9
 
 | |
1BZL
 
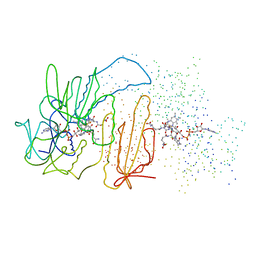 | | CRYSTAL STRUCTURE OF TRYPANOSOMA CRUZI TRYPANOTHIONE REDUCTASE IN COMPLEX WITH TRYPANOTHIONE, AND THE STRUCTURE-BASED DISCOVERY OF NEW NATURAL PRODUCT INHIBITORS | | Descriptor: | BIS(GAMMA-GLUTAMYL-CYSTEINYL-GLYCINYL)SPERMIDINE, FLAVIN-ADENINE DINUCLEOTIDE, TRYPANOTHIONE REDUCTASE (OXIDIZED FORM) | | Authors: | Bond, C.S, Zhang, Y, Berriman, M, Cunningham, M, Fairlamb, A.H, Hunter, W.N. | | Deposit date: | 1998-11-02 | | Release date: | 1999-11-10 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of Trypanosoma cruzi trypanothione reductase in complex with trypanothione, and the structure-based discovery of new natural product inhibitors.
Structure Fold.Des., 7, 1999
|
|
5EXC
 
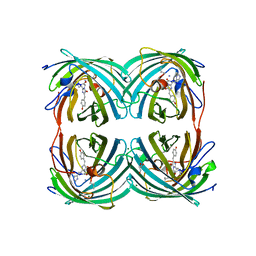 | |
5M8S
 
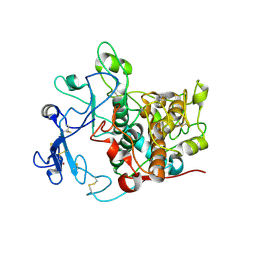 | | Crystal structure of human tyrosinase related protein 1 mutant (T391V-R374S-Y362F) in complex with phenylthiourea (PTU) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Lai, X, Soler-Lopez, M, Wichers, H.J, Dijkstra, B.W. | | Deposit date: | 2016-10-29 | | Release date: | 2018-01-31 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Phenylthiourea Binding to Human Tyrosinase-Related Protein 1
Int J Mol Sci, 2020
|
|
1C2A
 
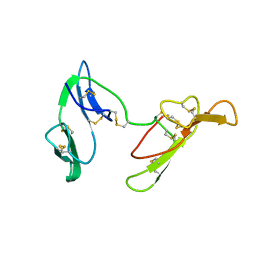 | | CRYSTAL STRUCTURE OF BARLEY BBI | | Descriptor: | BOWMAN-BIRK TRYPSIN INHIBITOR | | Authors: | Song, H.K, Kim, Y.S, Yang, J.K, Moon, J, Lee, J.Y, Suh, S.W. | | Deposit date: | 1999-07-23 | | Release date: | 1999-12-29 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of a 16 kDa double-headed Bowman-Birk trypsin inhibitor from barley seeds at 1.9 A resolution.
J.Mol.Biol., 293, 1999
|
|
1C3N
 
 | | CRYSTAL STRUCTURE OF HELIANTHUS TUBEROSUS LECTIN COMPLEXED TO MAN(1-2)MAN | | Descriptor: | AGGLUTININ, alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose | | Authors: | Bourne, Y, Zamboni, V, Barre, A, Peumans, W.J, van Damme, E.J.M, Rouge, P. | | Deposit date: | 1999-07-28 | | Release date: | 2000-01-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Helianthus tuberosus lectin reveals a widespread scaffold for mannose-binding lectins.
Structure Fold.Des., 7, 1999
|
|
7A64
 
 | |
1C1T
 
 | |
1BR2
 
 | | SMOOTH MUSCLE MYOSIN MOTOR DOMAIN COMPLEXED WITH MGADP.ALF4 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, MYOSIN, ... | | Authors: | Dominguez, R, Trybus, K.M, Cohen, C. | | Deposit date: | 1998-08-26 | | Release date: | 1998-09-09 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of a vertebrate smooth muscle myosin motor domain and its complex with the essential light chain: visualization of the pre-power stroke state.
Cell(Cambridge,Mass.), 94, 1998
|
|
1BMC
 
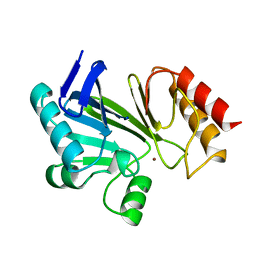 | | STRUCTURE OF A ZINC METALLO-BETA-LACTAMASE FROM BACILLUS CEREUS | | Descriptor: | METALLO-BETA-LACTAMASE, ZINC ION | | Authors: | Carfi, A, Pares, S, Duee, E, Dideberg, O. | | Deposit date: | 1995-06-16 | | Release date: | 1996-08-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The 3-D structure of a zinc metallo-beta-lactamase from Bacillus cereus reveals a new type of protein fold.
EMBO J., 14, 1995
|
|
1C2L
 
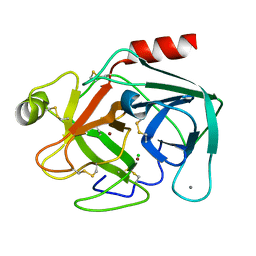 | |
1BMN
 
 | |
1C3F
 
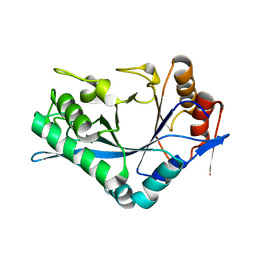 | | Endo-Beta-N-Acetylglucosaminidase H, D130N Mutant | | Descriptor: | ENDO-BETA-N-ACETYLGLUCOSAMINIDASE H | | Authors: | Rao, V, Cui, T, Guan, C, Van Roey, P. | | Deposit date: | 1999-07-27 | | Release date: | 1999-11-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Mutations of endo-beta-N-acetylglucosaminidase H active site residues Asp130 and Glu132: activities and conformations.
Protein Sci., 8, 1999
|
|
5F9F
 
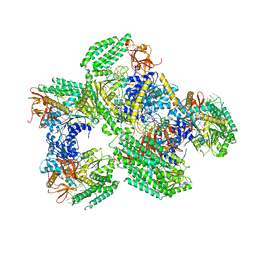 | | Crystal structure of RIG-I helicase-RD in complex with 24-mer blunt-end hairpin RNA | | Descriptor: | (R,R)-2,3-BUTANEDIOL, MAGNESIUM ION, Probable ATP-dependent RNA helicase DDX58, ... | | Authors: | Wang, C, Marcotrigiano, J, Miller, M.T, Jiang, F. | | Deposit date: | 2015-12-09 | | Release date: | 2016-01-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.601 Å) | | Cite: | Structural basis for m7G recognition and 2'-O-methyl discrimination in capped RNAs by the innate immune receptor RIG-I.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
1C3K
 
 | | CRYSTAL STRUCTURE OF HELIANTHUS TUBEROSUS LECTIN | | Descriptor: | AGGLUTININ | | Authors: | Bourne, Y, Zamboni, V, Barre, A, Peumans, W.J, van Damme, E.J.M, Rouge, P. | | Deposit date: | 1999-07-28 | | Release date: | 2000-01-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Helianthus tuberosus lectin reveals a widespread scaffold for mannose-binding lectins.
Structure Fold.Des., 7, 1999
|
|
1BPJ
 
 | | THYMIDYLATE SYNTHASE R178T, R179T DOUBLE MUTANT | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, POTASSIUM ION, PROTEIN (THYMIDYLATE SYNTHASE) | | Authors: | Morse, R.J, Finer-Moore, J.S, Stroud, R.M. | | Deposit date: | 1998-08-11 | | Release date: | 1998-08-19 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Energetic contributions of four arginines to phosphate-binding in thymidylate synthase are more than additive and depend on optimization of "effective charge balance".
Biochemistry, 39, 2000
|
|
7AK6
 
 | | Cryo-EM structure of ND6-P25L mutant respiratory complex I from Mus musculus at 3.8 A | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOETHANOLAMINE, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Yin, Z, Bridges, H.R, Grba, D, Hirst, J. | | Deposit date: | 2020-09-29 | | Release date: | 2021-02-03 | | Last modified: | 2021-02-10 | | Method: | ELECTRON MICROSCOPY (3.82 Å) | | Cite: | Structural basis for a complex I mutation that blocks pathological ROS production.
Nat Commun, 12, 2021
|
|
1BJK
 
 | | FERREDOXIN:NADP+ REDUCTASE MUTANT WITH ARG 264 REPLACED BY GLU (R264E) | | Descriptor: | FERREDOXIN--NADP+ REDUCTASE, FLAVIN-ADENINE DINUCLEOTIDE, SULFATE ION | | Authors: | Hermoso, J.A, Mayoral, T, Medina, M, Martinez-Ripoll, M, Martinez-Julvez, M, Sanz-Aparicio, J, Gomez-Moreno, C. | | Deposit date: | 1998-06-25 | | Release date: | 1998-11-04 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Role of Arg100 and Arg264 from Anabaena PCC 7119 ferredoxin-NADP+ reductase for optimal NADP+ binding and electron transfer.
Biochemistry, 37, 1998
|
|
