6LKN
 
 | | Crystal structure of ATP11C-CDC50A in PtdSer-bound E2P state | | Descriptor: | 1-deoxy-alpha-D-mannopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, Cell cycle control protein 50A, ... | | Authors: | Abe, K, Irie, K, Nakanishi, H, Hasegawa, K. | | Deposit date: | 2019-12-19 | | Release date: | 2020-06-10 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | Crystal structure of a human plasma membrane phospholipid flippase.
J.Biol.Chem., 295, 2020
|
|
1ONE
 
 | | YEAST ENOLASE COMPLEXED WITH AN EQUILIBRIUM MIXTURE OF 2'-PHOSPHOGLYCEATE AND PHOSPHOENOLPYRUVATE | | Descriptor: | 2-PHOSPHOGLYCERIC ACID, ENOLASE, MAGNESIUM ION, ... | | Authors: | Larsen, T.M, Wedekind, J.E, Rayment, I, Reed, G.H. | | Deposit date: | 1995-12-05 | | Release date: | 1997-01-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A carboxylate oxygen of the substrate bridges the magnesium ions at the active site of enolase: structure of the yeast enzyme complexed with the equilibrium mixture of 2-phosphoglycerate and phosphoenolpyruvate at 1.8 A resolution.
Biochemistry, 35, 1996
|
|
4MDG
 
 | | Closo Carborane Carbonic Anhydrase Inhibitor | | Descriptor: | 1-(sulfamoylamino)methyl-1,2-dicarba-closo-dodecaborane, Carbonic anhydrase 2, ZINC ION | | Authors: | Brynda, J, Mader, P, Rezacova, P. | | Deposit date: | 2013-08-22 | | Release date: | 2014-01-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Carborane-based carbonic anhydrase inhibitors.
Angew.Chem.Int.Ed.Engl., 52, 2013
|
|
2PPY
 
 | | Crystal structure of Enoyl-CoA hydrates (gk_1992) from Geobacillus Kaustophilus HTA426 | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Enoyl-CoA hydratase | | Authors: | Kanaujia, S.P, Jeyakanthan, J, Kavyashree, M, Sekar, K, Ebihara, A, Kuramitsu, S, Shinkai, A, Shiro, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-05-01 | | Release date: | 2008-05-06 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Crystal structure of Enoyl-CoA hydrates (gk_1992) from Geobacillus Kaustophilus HTA426
To be Published
|
|
4QFX
 
 | | Crystal structure of the tetrameric dGTP/dATP-bound SAMHD1 (RN206) mutant catalytic core | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, Deoxynucleoside triphosphate triphosphohydrolase SAMHD1, ... | | Authors: | Koharudin, L.M.I, Wu, Y, DeLucia, M, Mehrens, J, Gronenborn, A.M, Ahn, J. | | Deposit date: | 2014-05-21 | | Release date: | 2014-10-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Basis of Allosteric Activation of Sterile alpha Motif and Histidine-Aspartate Domain-containing Protein 1 (SAMHD1) by Nucleoside Triphosphates.
J.Biol.Chem., 289, 2014
|
|
6SEB
 
 | | Cold-adapted beta-D-galactosidase from Arthrobacter sp. 32cB in complex with IPTG | | Descriptor: | 1-methylethyl 1-thio-beta-D-galactopyranoside, ACETATE ION, Beta-galactosidase, ... | | Authors: | Rutkiewicz, M, Bujacz, A, Kaminska, P, Bujacz, G. | | Deposit date: | 2019-07-29 | | Release date: | 2019-09-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.272 Å) | | Cite: | Active Site Architecture and Reaction Mechanism Determination of Cold Adapted beta-d-galactosidase fromArthrobactersp. 32cB.
Int J Mol Sci, 20, 2019
|
|
4MAO
 
 | | RSK2 T493M C-Terminal Kinase Domain in Complex with RMM58 | | Descriptor: | (2Z)-2-(1H-1,2,4-triazol-1-yl)-3-[3-(3,4,5-trimethoxyphenyl)-1H-indazol-5-yl]prop-2-enenitrile, Ribosomal protein S6 kinase alpha-3, SODIUM ION | | Authors: | Miller, R.M, Taunton, J. | | Deposit date: | 2013-08-16 | | Release date: | 2014-10-22 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Design of reversible, cysteine-targeted Michael acceptors guided by kinetic and computational analysis.
J.Am.Chem.Soc., 136, 2014
|
|
1N97
 
 | | Crystal Structure of CYP175A1 from Thermus thermophillus strain HB27 | | Descriptor: | 1,2-ETHANEDIOL, CYP175A1, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Yano, J.K, Blasco, F, Li, H, Schmid, R.D, Henne, A, Poulos, T.L. | | Deposit date: | 2002-11-22 | | Release date: | 2003-02-25 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Preliminary Characterization and Crystal Structure of a Thermostable Cytochrome P450 from Thermus thermophilus
J.Biol.Chem., 278, 2003
|
|
8AAA
 
 | | Crystal structure of SARS-CoV-2 S RBD in complex with a stapled peptide | | Descriptor: | 1,1',1''-(1,3,5-triazinane-1,3,5-triyl)tripropan-1-one, Spike protein S1, Stapled peptide | | Authors: | Brear, P, Chen, L, Gaynor, K, Harman, M, Dods, R, Hyvonen, M. | | Deposit date: | 2022-06-30 | | Release date: | 2023-06-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Multivalent bicyclic peptides are an effective antiviral modality that can potently inhibit SARS-CoV-2.
Nat Commun, 14, 2023
|
|
2C6E
 
 | | Aurora A kinase activated mutant (T287D) in complex with a 5- aminopyrimidinyl quinazoline inhibitor | | Descriptor: | N-{5-[(7-{[(2S)-2-HYDROXY-3-PIPERIDIN-1-YLPROPYL]OXY}-6-METHOXYQUINAZOLIN-4-YL)AMINO]PYRIMIDIN-2-YL}BENZAMIDE, SERINE/THREONINE-PROTEIN KINASE 6 | | Authors: | Pauptit, R.A, Pannifer, A.D, Breed, J, McMiken, H.H.J, Rowsell, S, Anderson, M. | | Deposit date: | 2005-11-09 | | Release date: | 2006-01-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Sar and Inhibitor Complex Structure Determination of a Novel Class of Potent and Specific Aurora Kinase Inhibitors.
Bioorg.Med.Chem.Lett., 16, 2006
|
|
1UTG
 
 | | REFINEMENT OF THE C2221 CRYSTAL FORM OF OXIDIZED UTEROGLOBIN AT 1.34 ANGSTROMS RESOLUTION | | Descriptor: | UTEROGLOBIN | | Authors: | Morize, I, Surcouf, E, Vaney, M.C, Buehner, M, Mornon, J.P. | | Deposit date: | 1989-04-03 | | Release date: | 1989-10-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Refinement of the C222(1) crystal form of oxidized uteroglobin at 1.34 A resolution.
J.Mol.Biol., 194, 1987
|
|
2ZOU
 
 | | Crystal structure of human F-spondin reeler domain (fragment 2) | | Descriptor: | 1,2-ETHANEDIOL, Spondin-1 | | Authors: | Nagae, M, Nogi, T, Takagi, J. | | Deposit date: | 2008-06-07 | | Release date: | 2008-10-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure of the F-spondin reeler domain reveals a unique beta-sandwich fold with a deformable disulfide-bonded loop
Acta Crystallogr.,Sect.D, 64, 2008
|
|
4K2M
 
 | | Crystal structure of ntda from bacillus subtilis in complex with the plp external aldimine adduct with kanosamine-6-phosphate | | Descriptor: | 1,2-ETHANEDIOL, 3-deoxy-3-[(E)-({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene)amino]-6-O-phosphono-alpha-D-gluco pyranose, ACETATE ION, ... | | Authors: | Van Straaten, K.E, Palmer, D.R.J, Sanders, D.A.R. | | Deposit date: | 2013-04-09 | | Release date: | 2013-10-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | The Structure of NtdA, a Sugar Aminotransferase Involved in the Kanosamine Biosynthetic Pathway in Bacillus subtilis, Reveals a New Subclass of Aminotransferases.
J.Biol.Chem., 288, 2013
|
|
5BXN
 
 | | Yeast 20S proteasome beta2-G170A mutant in complex with Bortezomib | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, N-[(1R)-1-(DIHYDROXYBORYL)-3-METHYLBUTYL]-N-(PYRAZIN-2-YLCARBONYL)-L-PHENYLALANINAMIDE, ... | | Authors: | Huber, E.M, Groll, M. | | Deposit date: | 2015-06-09 | | Release date: | 2016-06-15 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Defective immuno- and thymoproteasome assembly causes severe immunodeficiency.
Sci Rep, 8, 2018
|
|
4BVG
 
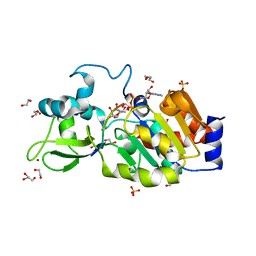 | | CRYSTAL STRUCTURE OF HUMAN SIRT3 IN COMPLEX WITH NATIVE ALKYLIMIDATE FORMED FROM ACETYL-LYSINE ACS2-PEPTIDE CRYSTALLIZED IN PRESENCE OF THE INHIBITOR EX-527 | | Descriptor: | (2R,3R,4S,5R)-5-({[(R)-{[(R)-{[(2R,3S,4R,5R)-5-(6-AMINO-9H-PURIN-9-YL)-3,4-DIHYDROXYTETRAHYDROFURAN-2-YL]METHOXY}(HYDROXY)PHOSPHORYL]OXY}(HYDROXY)PHOSPHORYL]OXY}METHYL)-3,4-DIHYDROXYTETRAHYDROFURAN-2-YL ACETATE, 1,2-ETHANEDIOL, ACETYL-COENZYME A SYNTHETASE 2-LIKE, ... | | Authors: | Nguyen, G.T.T, Gertz, M, Weyand, M, Steegborn, C. | | Deposit date: | 2013-06-25 | | Release date: | 2013-07-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Ex-527 inhibits Sirtuins by exploiting their unique NAD+-dependent deacetylation mechanism.
Proc. Natl. Acad. Sci. U.S.A., 110, 2013
|
|
3OQ8
 
 | |
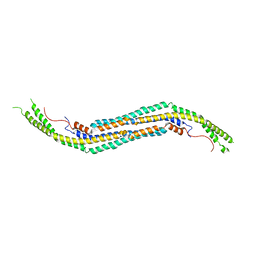
3QNI
 
 | |
2RAZ
 
 | |
3ZJD
 
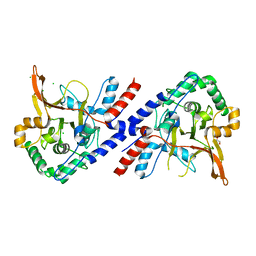 | | A20 OTU domain in reduced, active state at 1.87 A resolution | | Descriptor: | 1,2-ETHANEDIOL, A20P50, CHLORIDE ION | | Authors: | Kulathu, Y, Garcia, F.J, Mevissen, T.E.T, Busch, M, Arnaudo, N, Carroll, K.S, Barford, D, Komander, D. | | Deposit date: | 2013-01-17 | | Release date: | 2013-03-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Regulation of A20 and Other Otu Deubiquitinases by Reversible Oxidation
Nat.Commun., 4, 2013
|
|
8A7N
 
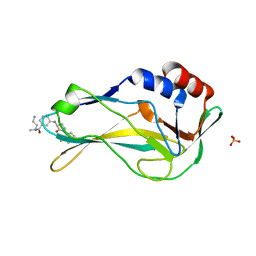 | | Crystal Structure of human Brachyury G177D variant in complex with (S)-N-(3-aminopropyl)-3-((1-(2-fluorophenyl)-2-oxopyrrolidin-3-yl)amino)-N-methylbenzamide (CF-2-125) | | Descriptor: | N-(3-azanylpropyl)-3-[[(3S)-1-(2-fluorophenyl)-2-oxidanylidene-pyrrolidin-3-yl]amino]-N-methyl-benzamide, PHOSPHATE ION, T-box transcription factor T | | Authors: | Newman, J.A, Gavard, A, Aitkenhead, H, Imprachim, N, Sherestha, L, Burgess-Brown, N.A, von Delft, F, Bountra, C, Gileadi, O. | | Deposit date: | 2022-06-21 | | Release date: | 2022-10-05 | | Last modified: | 2025-02-26 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural insights into human brachyury DNA recognition and discovery of progressible binders for cancer therapy.
Nat Commun, 16, 2025
|
|
3Q90
 
 | | Crystal structure of the NTF2 domain of Ras GTPase-activating protein-binding protein 1 | | Descriptor: | Ras GTPase-activating protein-binding protein 1 | | Authors: | Welin, M, Tresaugues, L, Arrowsmith, C.H, Berglund, H, Bountra, C, Collins, R, Edwards, A.M, Ekblad, T, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, I, Karlberg, T, Kol, S, Kotenyova, T, Kouznetsova, E, Moche, M, Nyman, T, Persson, C, Schuler, H, Schutz, P, Siponen, M.I, Thorsell, A.G, Van Der Berg, S, Wahlberg, E, Weigelt, J, Nordlund, P, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-01-07 | | Release date: | 2011-02-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of the NTF2 domain of Ras GTPase-activating protein-binding protein 1
To be Published
|
|
3QA2
 
 | | X-ray Structure of ketohexokinase in complex with a pyrimidopyrimidine analog 2 | | Descriptor: | Ketohexokinase, N~8~-(cyclopropylmethyl)-N~4~-(2-methylphenyl)-2-(piperazin-1-yl)pyrimido[5,4-d]pyrimidine-4,8-diamine, SULFATE ION | | Authors: | Abad, M.C. | | Deposit date: | 2011-01-10 | | Release date: | 2012-01-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.519 Å) | | Cite: | Inhibitors of Ketohexokinase: Discovery of Pyrimidinopyrimidines with Specific Substitution that Complements the ATP-Binding Site.
ACS Med Chem Lett, 2, 2011
|
|
3CEJ
 
 | | Human glycogen phosphorylase (tense state) in complex with the allosteric inhibitor AVE2865 | | Descriptor: | 1-{2-[3-(2-Chloro-4,5-difluoro-benzoyl)-ureido]-4-fluoro-phenyl}-piperidine-4-carboxylic acid, Glycogen phosphorylase, liver form, ... | | Authors: | Wendt, K.U, Dreyer, M.K, Anderka, O, Klabunde, T, Loenze, P, Defossa, E, Schmoll, D. | | Deposit date: | 2008-02-29 | | Release date: | 2008-05-27 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Thermodynamic characterization of allosteric glycogen phosphorylase inhibitors.
Biochemistry, 47, 2008
|
|
5RHD
 
 | | PanDDA analysis group deposition SARS-CoV-2 main protease fragment screen -- Crystal Structure of SARS-CoV-2 main protease in complex with SF013 (Mpro-x2193) | | Descriptor: | 1-[4-(methylsulfonyl)phenyl]piperazine, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Owen, C.D, Douangamath, A, Lukacik, P, Powell, A.J, Strain-Damerell, C.M, Resnick, E, Krojer, T, Gehrtz, P, Wild, C, Aimon, A, Brandao-Neto, J, Carbery, A, Dunnett, L, Skyner, R, Snee, M, Keeley, A, Keseru, G.M, London, N, Walsh, M.A, von Delft, F. | | Deposit date: | 2020-05-16 | | Release date: | 2020-06-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Crystallographic and electrophilic fragment screening of the SARS-CoV-2 main protease.
Nat Commun, 11, 2020
|
|
4CHY
 
 | | Interrogating HIV integrase for compounds that bind- a SAMPL challenge | | Descriptor: | 1,2-ETHANEDIOL, 5-(1H-indol-3-ylmethyl)-1,3-benzodioxole-4-carboxylic acid, ACETIC ACID, ... | | Authors: | Peat, T.S. | | Deposit date: | 2013-12-04 | | Release date: | 2013-12-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Interrogating HIV Integrase for Compounds that Bind- a Sampl Challenge.
J.Comput.Aided Mol.Des., 28, 2014
|
|
