7LG7
 
 | | Crystal structure of CoV-2 Nsp3 Macrodomain complex with PARG345 | | Descriptor: | 3-[(1,3-dimethyl-2,6-dioxo-2,3,6,9-tetrahydro-1H-purin-8-yl)sulfanyl]-N-{[2-(morpholin-4-yl)ethyl]sulfonyl}propanamide, Non-structural protein 3, SULFATE ION | | Authors: | Arvai, A, Brosey, C.A, Bommagani, S, Link, T, Jones, D.E, Ahmed, Z, Tainer, J.A. | | Deposit date: | 2021-01-19 | | Release date: | 2021-02-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Targeting SARS-CoV-2 Nsp3 macrodomain structure with insights from human poly(ADP-ribose) glycohydrolase (PARG) structures with inhibitors.
Prog.Biophys.Mol.Biol., 163, 2021
|
|
4Z9D
 
 | | EcPltA | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Pertussis toxin-like subunit ArtA, SODIUM ION | | Authors: | Littler, D.R, Johnson, M.D, Summers, R.J, Schembri, M.A, Rossjohn, J, Beddoe, T. | | Deposit date: | 2015-04-10 | | Release date: | 2016-10-12 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure and function analyses of a pertussis-like toxin from pathogenic Escherichia coli reveal a distinct mechanism of inhibition of trimeric G proteins.
J. Biol. Chem., 2017
|
|
7LKA
 
 | | Crystal structure of SARS-CoV-2 RBD-targeting antibody COV107-23 | | Descriptor: | ACETATE ION, COV107-23 heavy chain, COV107-23 light chain | | Authors: | Yuan, M, Zhu, X, Wilson, I.A, Wu, N.C. | | Deposit date: | 2021-02-02 | | Release date: | 2021-02-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.981 Å) | | Cite: | Sequence signatures of two public antibody clonotypes that bind SARS-CoV-2 receptor binding domain.
Nat Commun, 12, 2021
|
|
7LK9
 
 | | Crystal structure of SARS-CoV-2 RBD-targeting antibody COV107-23 HC + COVD21-C8 LC | | Descriptor: | COV107-23 heavy chain, COVD21-C8 light chain | | Authors: | Yuan, M, Zhu, X, Wilson, I.A, Wu, N.C. | | Deposit date: | 2021-02-01 | | Release date: | 2021-02-17 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Sequence signatures of two public antibody clonotypes that bind SARS-CoV-2 receptor binding domain.
Nat Commun, 12, 2021
|
|
6PD1
 
 | |
3LP9
 
 | | Crystal structure of LS24, A Seed Albumin from Lathyrus sativus | | Descriptor: | CALCIUM ION, CHLORIDE ION, LS-24, ... | | Authors: | Gaur, V, Qureshi, I.A, Singh, A, Chanana, V, Salunke, D.M. | | Deposit date: | 2010-02-05 | | Release date: | 2010-02-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure and functional insights of hemopexin fold protein from grass pea
Plant Physiol., 152, 2010
|
|
5NG2
 
 | | Structure of RIP2K(D146N) with bound Staurosporine | | Descriptor: | PHOSPHATE ION, Receptor-interacting serine/threonine-protein kinase 2, STAUROSPORINE | | Authors: | Pellegrini, E, Cusack, S. | | Deposit date: | 2017-03-16 | | Release date: | 2017-06-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structures of the inactive and active states of RIP2 kinase inform on the mechanism of activation.
PLoS ONE, 12, 2017
|
|
6S50
 
 | |
4R0W
 
 | |
7ZU6
 
 | | Polyoxidovanadate interaction with proteins: crystal structure of lysozyme bound to tetra-vanadate ion (structure 1) | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CYCLO-TETRAMETAVANADATE, Lysozyme, ... | | Authors: | Tito, G, Merlino, A, Ferraro, G. | | Deposit date: | 2022-05-11 | | Release date: | 2023-05-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.183 Å) | | Cite: | Stabilization and Binding of [V 4 O 12 ] 4- and Unprecedented [V 20 O 54 (NO 3 )] n- to Lysozyme upon Loss of Ligands and Oxidation of the Potential Drug V IV O(acetylacetonato) 2.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
3E9U
 
 | | Crystal structure of Calx CBD2 domain | | Descriptor: | Na/Ca exchange protein | | Authors: | Wu, M, Zheng, L. | | Deposit date: | 2008-08-23 | | Release date: | 2009-01-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of CBD2 from the Drosophila Na(+)/Ca(2+) exchanger: diversity of Ca(2+) regulation and its alternative splicing modification.
J.Mol.Biol., 387, 2009
|
|
5NHK
 
 | | Structure of Ferric uptake regulator from francisella tularensis with Iron | | Descriptor: | FE (III) ION, Ferric uptake regulation protein, ZINC ION | | Authors: | Perard, J, Arnaud, L, Carpentier, P, Michaud-Soret, I, Cavazza, C. | | Deposit date: | 2017-03-21 | | Release date: | 2018-05-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and functional studies of the metalloregulator Fur identify a promoter-binding mechanism and its role inFrancisella tularensisvirulence.
Commun Biol, 1, 2018
|
|
5X4F
 
 | |
6PDK
 
 | |
6V12
 
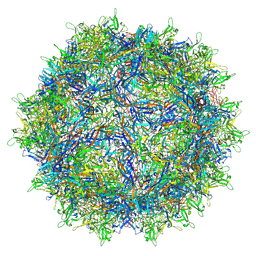 | | Empty AAV8 particles | | Descriptor: | Capsid protein | | Authors: | Mietzsch, M, Agbandje-McKenna, M. | | Deposit date: | 2019-11-19 | | Release date: | 2019-12-11 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (3.08 Å) | | Cite: | Comparative Analysis of the Capsid Structures of AAVrh.10, AAVrh.39, and AAV8.
J.Virol., 94, 2020
|
|
6IK7
 
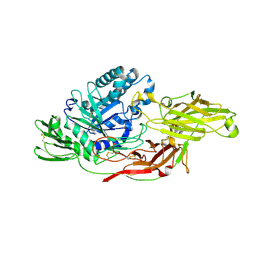 | | Crystal structure of tomato beta-galactosidase (TBG) 4 in complex with beta-1,3-galactobiose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-galactosidase, ... | | Authors: | Matsuyama, K, Nakae, S, Igarashi, K, Tada, T, Ishimaru, M. | | Deposit date: | 2018-10-15 | | Release date: | 2018-11-28 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Substrate-recognition mechanism of tomato beta-galactosidase 4 using X-ray crystallography and docking simulation.
Planta, 252, 2020
|
|
6PFM
 
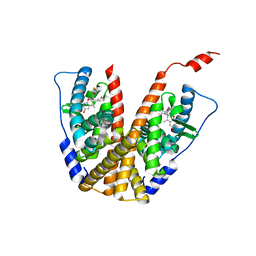 | | Crystal structure of GDC-0927 bound to estrogen receptor alpha | | Descriptor: | (2S)-2-(4-{2-[3-(fluoromethyl)azetidin-1-yl]ethoxy}phenyl)-3-(3-hydroxyphenyl)-4-methyl-2H-1-benzopyran-6-ol, Estrogen receptor | | Authors: | Kiefer, J.R, Vinogradova, M, Liang, J, Zhang, B, Wang, X, Zbieg, J.R, Labadie, S.S, Li, J, Ray, N.C, Ortwine, D. | | Deposit date: | 2019-06-21 | | Release date: | 2019-07-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Discovery of a C-8 hydroxychromene as a potent degrader of estrogen receptor alpha with improved rat oral exposure over GDC-0927.
Bioorg.Med.Chem.Lett., 29, 2019
|
|
6UPX
 
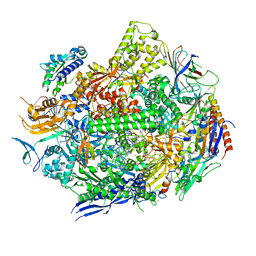 | | RNA polymerase II elongation complex with 5-guanidinohydantoin lesion in state 1 | | Descriptor: | DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB11, DNA-directed RNA polymerase II subunit RPB2, ... | | Authors: | Oh, J, Wang, D. | | Deposit date: | 2019-10-18 | | Release date: | 2020-06-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | RNA polymerase II stalls on oxidative DNA damage via a torsion-latch mechanism involving lone pair-pi and CH-pi interactions.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
7WK8
 
 | |
6ILH
 
 | | Crystal Structure of human lysyl-tRNA synthetase L350H mutant | | Descriptor: | 5'-O-[(L-LYSYLAMINO)SULFONYL]ADENOSINE, Lysine-tRNA ligase | | Authors: | Hei, Z, Liu, Z, Wang, J, Fang, P. | | Deposit date: | 2018-10-18 | | Release date: | 2019-02-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | Retractile lysyl-tRNA synthetase-AIMP2 assembly in the human multi-aminoacyl-tRNA synthetase complex.
J. Biol. Chem., 294, 2019
|
|
5H35
 
 | | Crystal structures of the TRIC trimeric intracellular cation channel orthologue from Sulfolobus solfataricus | | Descriptor: | 1,2-DIMYRISTOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Fab Heavy Chain, Fab Light Chain, ... | | Authors: | Kasuya, G, Hiraizumi, M, Hattori, M, Nureki, O. | | Deposit date: | 2016-10-20 | | Release date: | 2017-01-11 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.642 Å) | | Cite: | Crystal structures of the TRIC trimeric intracellular cation channel orthologues
Cell Res., 26, 2016
|
|
5UII
 
 | |
7VW5
 
 | |
6M09
 
 | |
6PFQ
 
 | |
