2L5K
 
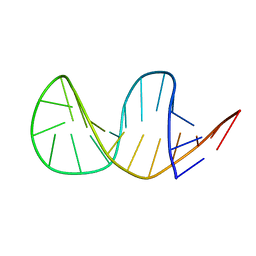 | | Solution structure of truncated 23-mer DNA MUC1 aptamer | | Descriptor: | DNA (5'-R(*(N68)P*G)-D(*CP*AP*GP*TP*TP*GP*AP*TP*CP*CP*TP*TP*TP*GP*GP*AP*TP*AP*CP*CP*CP*TP*GP*GP*T)-3') | | Authors: | Cognet, J, Baouendi, M, Hantz, E, Missailidis, S, Herve du Penhoat, C, Piotto, M. | | Deposit date: | 2010-11-02 | | Release date: | 2011-12-07 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a truncated anti-MUC1 DNA aptamer determined by mesoscale modeling and NMR.
Febs J., 279, 2012
|
|
6U3R
 
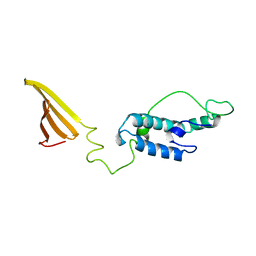 | |
2LGM
 
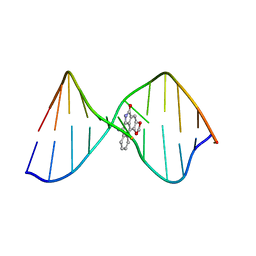 | | Structure of DNA Containing an Aristolactam II-dA Lesion | | Descriptor: | DNA (5'-D(*CP*GP*TP*AP*CP*AP*CP*AP*TP*GP*C)-3'), DNA (5'-D(*GP*CP*AP*TP*GP*TP*GP*TP*AP*CP*G)-3'), [1,3]benzodioxolo[6,5,4-cd]benzo[f]indol-5(6H)-one | | Authors: | Lukin, M, Zaliznyak, T, Johnson, F, de los Santos, C. | | Deposit date: | 2011-07-28 | | Release date: | 2011-11-30 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure and stability of DNA containing an aristolactam II-dA lesion: implications for the NER recognition of bulky adducts.
Nucleic Acids Res., 40, 2012
|
|
2LWO
 
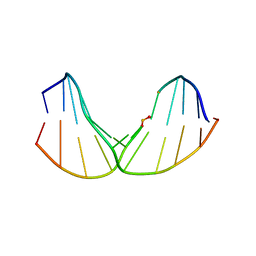 | | Solution Structure of Duplex DNA Containing a b-Carba-Fapy-dG Lesion | | Descriptor: | DNA (5'-D(*G*CP*GP*TP*AP*C*(LWM)P*CP*AP*TP*GP*C)-3'), DNA (5'-D(*GP*CP*AP*TP*GP*CP*GP*TP*AP*CP*G)-3') | | Authors: | Zalianyak, T, de los Santos, C, Lukin, M, Attaluri, S, Johnson, F. | | Deposit date: | 2012-08-03 | | Release date: | 2013-02-20 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of Duplex DNA Containing a b-Carba-Fapy-dG Lesion
Chem.Res.Toxicol., 25, 2012
|
|
2LWN
 
 | | Solution Structure of Duplex DNA Containing a b-Carba-Fapy-dG Lesion | | Descriptor: | DNA (5'-D(*CP*GP*TP*AP*C*(LWM)P*CP*AP*TP*GP*C)-3'), DNA (5'-D(*GP*CP*AP*TP*GP*CP*GP*TP*AP*CP*G)-3') | | Authors: | Zalianyak, T, de los Santos, C, Lukin, M, Attaluri, S, Johnson, F. | | Deposit date: | 2012-08-03 | | Release date: | 2013-02-20 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of Duplex DNA Containing a b-Carba-Fapy-dG Lesion
Chem.Res.Toxicol., 25, 2012
|
|
2LWM
 
 | | Solution Structure of Duplex DNA Containing a b-Carba-Fapy-dG Lesion | | Descriptor: | DNA (5'-D(*CP*GP*TP*AP*C*(LWM)P*CP*AP*TP*GP*C)-3'), DNA (5'-D(*GP*CP*AP*TP*GP*CP*GP*TP*AP*CP*G)-3') | | Authors: | Zalianyak, T, de los Santos, C, Lukin, M, Attaluri, S, Johnson, F. | | Deposit date: | 2012-08-03 | | Release date: | 2013-02-20 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of Duplex DNA Containing a b-Carba-Fapy-dG Lesion
Chem.Res.Toxicol., 25, 2012
|
|
2LT0
 
 | | NMR structure of duplex DNA containing the beta-OH-PdG dA base pair: A mutagenic intermediate of acrolein | | Descriptor: | DNA (5'-D(*CP*GP*TP*AP*CP*(63G)P*CP*AP*TP*GP*C)-3'), DNA (5'-D(*GP*CP*AP*TP*GP*AP*GP*TP*AP*CP*G)-3') | | Authors: | Zaliznyak, T, de los Santos, C, Lukin, M, El-khateeb, M, Bonala, R, Johnson, F. | | Deposit date: | 2012-05-09 | | Release date: | 2012-06-13 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | NMR structure of duplex DNA containing the alpha-OH-PdG.dA base pair: a mutagenic intermediate of acrolein.
Biopolymers, 93, 2010
|
|
2LSZ
 
 | | NMR structure of duplex DNA containing the alpha-OH-PdG dA base pair: A mutagenic intermediate of acrolein | | Descriptor: | DNA (5'-D(*CP*GP*TP*AP*CP*(63H)P*CP*AP*TP*GP*C)-3'), DNA (5'-D(*GP*CP*AP*TP*GP*AP*GP*TP*AP*CP*G)-3') | | Authors: | Zaliznyak, T, de los Santos, C, Lukin, M, El-khateeb, M, Bonala, R, Johnson, F. | | Deposit date: | 2012-05-09 | | Release date: | 2012-06-13 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | NMR structure of duplex DNA containing the alpha-OH-PdG.dA base pair: a mutagenic intermediate of acrolein.
Biopolymers, 93, 2010
|
|
2KE8
 
 | | NMR solution structure of metal-modified DNA | | Descriptor: | DNA (5'-D(*TP*TP*AP*AP*TP*TP*TP*(D33)P*(D33)P*(D33)P*AP*AP*AP*TP*TP*AP*A)-3'), SILVER ION | | Authors: | Johannsen, S, Duepre, N, Boehme, D, Mueller, J, Sigel, R.K.O. | | Deposit date: | 2009-01-27 | | Release date: | 2010-02-09 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a DNA double helix with consecutive metal-mediated base pairs.
Nat.Chem., 2, 2010
|
|
2O4Y
 
 | |
2NEO
 
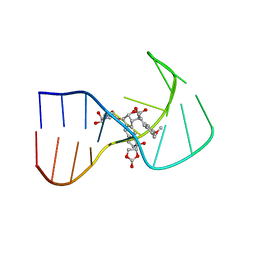 | | SOLUTION NMR STRUCTURE OF A TWO-BASE DNA BULGE COMPLEXED WITH AN ENEDIYNE CLEAVING ANALOG, 11 STRUCTURES | | Descriptor: | DNA (5'-D(*CP*CP*CP*GP*AP*TP*GP*CP*PGE*GP*CP*AP*AP*TP*TP*CP*GP*GP*G)-3'), SPIRO[[7-METHOXY-5-METHYL-1,2-DIHYDRO-NAPHTHALENE]-3,1'-[5-HYDROXY-9-[2-METHYLAMINO-2,6-DIDEOXYGALACTOPYRANOSYL-OXY]-5-(2-OXO-[1,3]DIOXOLAN-4-YL)-3A,5,9,9A-TETRAHYDRO-3H-1-OXA-CYCLOPENTA[A]-S-INDACEN-2-ONE]] | | Authors: | Stassinopoulos, A, Ji, J, Gao, X, Goldberg, I.H. | | Deposit date: | 1997-06-19 | | Release date: | 1998-01-28 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a two-base DNA bulge complexed with an enediyne cleaving analog.
Science, 272, 1996
|
|
2KXE
 
 | |
5K8B
 
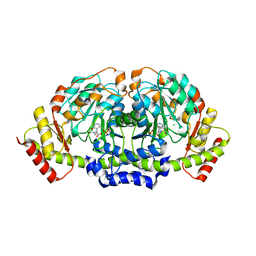 | | X-ray structure of KdnA, 8-amino-3,8-dideoxy-alpha-D-manno-octulosonate transaminase, from Shewanella oneidensis in the presence of the external aldimine with PLP and glutamate | | Descriptor: | 8-amino-3,8-dideoxy-alpha-D-manno-octulosonate transaminase, CHLORIDE ION, N-({3-HYDROXY-2-METHYL-5-[(PHOSPHONOOXY)METHYL]PYRIDIN-4-YL}METHYL)-D-GLUTAMIC ACID, ... | | Authors: | Holden, H.M, Thoden, J.B, Zachman-Brockmeyer, T.R. | | Deposit date: | 2016-05-28 | | Release date: | 2016-06-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structures of KdnB and KdnA from Shewanella oneidensis: Key Enzymes in the Formation of 8-Amino-3,8-Dideoxy-d-Manno-Octulosonic Acid.
Biochemistry, 55, 2016
|
|
7QBY
 
 | |
6U3S
 
 | |
7QIL
 
 | |
6WJG
 
 | | PKA RIIbeta holoenzyme with DnaJB1-PKAc fusion in fibrolamellar hepatoceullar carcinoma | | Descriptor: | DnaJ homolog subfamily B member 1, cAMP-dependent protein kinase catalytic subunit alpha fusion, cAMP-dependent protein kinase type II-beta regulatory subunit | | Authors: | Lu, T.-W, Aoto, P.C, Weng, J.-H, Nielsen, C, Cash, J.N, Hall, J, Zhang, P, Simon, S.M, Cianfrocco, M.A, Taylor, S.S. | | Deposit date: | 2020-04-13 | | Release date: | 2020-12-02 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (6.2 Å) | | Cite: | Structural analyses of the PKA RII beta holoenzyme containing the oncogenic DnaJB1-PKAc fusion protein reveal protomer asymmetry and fusion-induced allosteric perturbations in fibrolamellar hepatocellular carcinoma.
Plos Biol., 18, 2020
|
|
6WJF
 
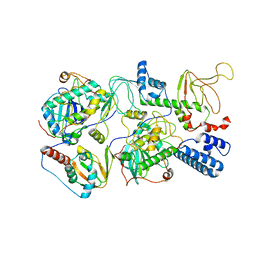 | | PKA RIIbeta holoenzyme with DnaJB1-PKAc fusion in fibrolamellar hepatoceullar carcinoma | | Descriptor: | DnaJ homolog subfamily B member 1,cAMP-dependent protein kinase catalytic subunit alpha fusion, cAMP-dependent protein kinase type II-beta regulatory subunit | | Authors: | Lu, T.-W, Aoto, P.C, Weng, J.-H, Nielsen, C, Cash, J.N, Hall, J, Zhang, P, Simon, S.M, Cianfrocco, M.A, Taylor, S.S. | | Deposit date: | 2020-04-13 | | Release date: | 2020-12-02 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (7.5 Å) | | Cite: | Structural analyses of the PKA RII beta holoenzyme containing the oncogenic DnaJB1-PKAc fusion protein reveal protomer asymmetry and fusion-induced allosteric perturbations in fibrolamellar hepatocellular carcinoma.
Plos Biol., 18, 2020
|
|
4EZU
 
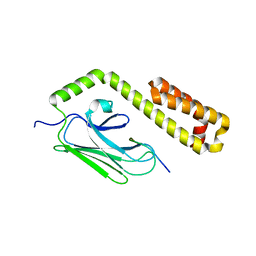 | |
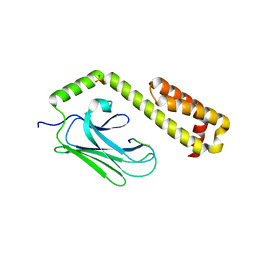
4EZS
 
 | |
7JSQ
 
 | | Refined structure of the C-terminal domain of DNAJB6b | | Descriptor: | DnaJ homolog subfamily B member 6 | | Authors: | Karamanos, T.K, Clore, G.M. | | Deposit date: | 2020-08-15 | | Release date: | 2020-12-02 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | An S/T motif controls reversible oligomerization of the Hsp40 chaperone DNAJB6b through subtle reorganization of a beta sheet backbone.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
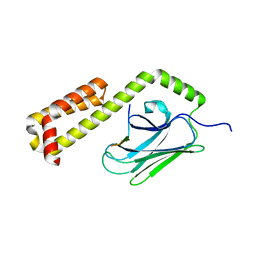
4EZV
 
 | |
2GSJ
 
 | | cDNA cloning and 1.75A crystal structure determination of PPL2, a novel chimerolectin from Parkia platycephala seeds exhibiting endochitinolytic activity | | Descriptor: | SULFATE ION, protein PPL-2 | | Authors: | Cavada, B.S, Moreno, F.B, da Rocha, B.A, de Azevedo Jr, W.F, Castellon, R.E.R, Goersch, G.V, Nagano, C.S, de Souza, E.P, Nascimento, K.S, Radis-Baptista, G, Delatorre, P, Leroy, Y, Toyama, M.H, Pinto, V.P, Sampaio, A.H, Barettino, D, Debray, H, Calvete, J.J, Sanz, L. | | Deposit date: | 2006-04-26 | | Release date: | 2007-03-13 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | cDNA cloning and 1.75 A crystal structure determination of PPL2, an endochitinase and N-acetylglucosamine-binding hemagglutinin from Parkia platycephala seeds
Febs J., 273, 2006
|
|
3QNJ
 
 | |
2M6Y
 
 | | The solution structure of the J-domain of human DnaJA1 | | Descriptor: | DnaJ homolog subfamily A member 1 | | Authors: | Stark, J.L, Mehla, K, Chaika, N, Acton, T.B, Xiao, R, Singh, P.K, Montelione, G.T, Powers, R, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2013-04-14 | | Release date: | 2013-06-26 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure and function of human DnaJ homologue subfamily a member 1 (DNAJA1) and its relationship to pancreatic cancer.
Biochemistry, 53, 2014
|
|
