2BU5
 
 | | crystal structures of human pyruvate dehydrogenase kinase 2 containing physiological and synthetic ligands | | Descriptor: | 4-({(2R,5S)-2,5-DIMETHYL-4-[(2R)-3,3,3-TRIFLUORO-2-HYDROXY-2-METHYLPROPANOYL]PIPERAZIN-1-YL}CARBONYL)BENZONITRILE, PYRUVATE DEHYDROGENASE KINASE ISOENZYME 2 | | Authors: | Knoechel, T.R, Tucker, A.D, Robinson, C.M, Phillips, C, Taylor, W, Bungay, P.J, Kasten, S.A, Roche, T.E, Brown, D.G. | | Deposit date: | 2005-06-08 | | Release date: | 2006-02-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Regulatory Roles of the N-Terminal Domain Based on Crystal Structures of Human Pyruvate Dehydrogenase Kinase 2 Containing Physiological and Synthetic Ligands.
Biochemistry, 45, 2006
|
|
4MPC
 
 | | Crystal structure of pyruvate dehydrogenase kinase isoform 2 in complex with inhibitor PS2 | | Descriptor: | 4-(isoindolin-2-ylsulfonyl)benzene-1,3-diol, L(+)-TARTARIC ACID, [Pyruvate dehydrogenase [lipoamide]] kinase isozyme 2, ... | | Authors: | Gui, W.J, Tso, S.C, Chuang, J.L, Wu, C.Y, Qi, X, Tambar, U.K, Wynn, R.M, Chuang, D.T. | | Deposit date: | 2013-09-12 | | Release date: | 2014-01-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.699 Å) | | Cite: | Structure-guided Development of Specific Pyruvate Dehydrogenase Kinase Inhibitors Targeting the ATP-binding Pocket.
J.Biol.Chem., 289, 2014
|
|
2BTZ
 
 | | crystal structures of human pyruvate dehydrogenase kinase 2 containing physiological and synthetic ligands | | Descriptor: | PYRUVATE DEHYDROGENASE KINASE ISOENZYME 2 | | Authors: | Knoechel, T.R, Tucker, A.D, Robinson, C.M, Phillips, C, Taylor, W, Bungay, P.J, Kasten, S.A, Roche, T.E, Brown, D.G. | | Deposit date: | 2005-06-08 | | Release date: | 2006-02-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Regulatory Roles of the N-Terminal Domain Based on Crystal Structures of Human Pyruvate Dehydrogenase Kinase 2 Containing Physiological and Synthetic Ligands.
Biochemistry, 45, 2006
|
|
2BEU
 
 | | Reactivity modulation of human branched-chain alpha-ketoacid dehydrogenase by an internal molecular switch | | Descriptor: | 2-OXOISOVALERATE DEHYDROGENASE ALPHA SUBUNIT, 2-OXOISOVALERATE DEHYDROGENASE BETA SUBUNIT, C2-1-HYDROXY-3-METHYL-PROPYL-THIAMIN DIPHOSPHATE, ... | | Authors: | Machius, M, Wynn, R.M, Chuang, J.L, Tomchick, D.R, Brautigam, C.A, Chuang, D.T. | | Deposit date: | 2004-11-30 | | Release date: | 2006-02-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | A Versatile Conformational Switch Regulates Reactivity in Human Branched-Chain Alpha-Ketoacid Dehydrogenase.
Structure, 14, 2006
|
|
2BEW
 
 | | Reactivity modulation of human branched-chain alpha-ketoacid dehydrogenase by an internal molecular switch | | Descriptor: | 2-OXOISOVALERATE DEHYDROGENASE ALPHA SUBUNIT, 2-OXOISOVALERATE DEHYDROGENASE BETA SUBUNIT, C2-1-HYDROXYPHENYL-THIAMIN DIPHOSPHATE, ... | | Authors: | Machius, M, Wynn, R.M, Chuang, J.L, Tomchick, D.R, Brautigam, C.A, Chuang, D.T. | | Deposit date: | 2004-11-30 | | Release date: | 2006-02-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | A Versatile Conformational Switch Regulates Reactivity in Human Branched-Chain Alpha-Ketoacid Dehydrogenase.
Structure, 14, 2006
|
|
7ZFW
 
 | |
2ZDX
 
 | | Inhibitor-bound structures of human pyruvate dehydrogenase kinase 4 | | Descriptor: | 4-[4-(4-methoxyphenyl)-5-methyl-1H-pyrazol-3-yl]benzene-1,3-diol, Pyruvate dehydrogenase kinase isozyme 4 | | Authors: | Kawamoto, M, Shiromizu, I, Kukimoto-niino, M, Tokmakov, A, Terada, T, Shirouzu, M, Matsusue, T, Yokoyama, S. | | Deposit date: | 2007-11-30 | | Release date: | 2008-12-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Inhibitor-bound structures of human pyruvate dehydrogenase kinase 4.
Acta Crystallogr.,Sect.D, 67, 2011
|
|
2E0A
 
 | | Crystal structure of human pyruvate dehydrogenase kinase 4 in complex with AMPPNP | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Pyruvate dehydrogenase kinase isozyme 4 | | Authors: | Kukimoto-Niino, M, Tokmakov, A, Terada, T, Shiromizu, I, Kawamoto, M, Matsusue, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-10-03 | | Release date: | 2007-10-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Inhibitor-bound structures of human pyruvate dehydrogenase kinase 4.
Acta Crystallogr.,Sect.D, 67, 2011
|
|
2E76
 
 | | Crystal Structure of the Cytochrome b6f Complex with tridecyl-stigmatellin (TDS) from M.laminosus | | Descriptor: | (7R,17E)-4-HYDROXY-N,N,N,7-TETRAMETHYL-7-[(8E)-OCTADEC-8-ENOYLOXY]-10-OXO-3,5,9-TRIOXA-4-PHOSPHAHEPTACOS-17-EN-1-AMINIUM 4-OXIDE, 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 8-HYDROXY-5,7-DIMETHOXY-3-METHYL-2-TRIDECYL-4H-CHROMEN-4-ONE, ... | | Authors: | Cramer, W.A, Yamashita, E, Zhang, H. | | Deposit date: | 2007-01-05 | | Release date: | 2007-06-12 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3.41 Å) | | Cite: | Structure of the Cytochrome b(6)f Complex: Quinone Analogue Inhibitors as Ligands of Heme c(n)
J.Mol.Biol., 370, 2007
|
|
7A5V
 
 | | CryoEM structure of a human gamma-aminobutyric acid receptor, the GABA(A)R-beta3 homopentamer, in complex with histamine and megabody Mb25 in lipid nanodisc | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Nakane, T, Kotecha, A, Sente, A, Yamashita, K, McMullan, G, Masiulis, S, Brown, P.M.G.E, Grigoras, I.T, Malinauskaite, L, Malinauskas, T, Miehling, J, Yu, L, Karia, D, Pechnikova, E.V, de Jong, E, Keizer, J, Bischoff, M, McCormack, J, Tiemeijer, P, Hardwick, S.W, Chirgadze, D.Y, Murshudov, G, Aricescu, A.R, Scheres, S.H.W. | | Deposit date: | 2020-08-22 | | Release date: | 2020-11-18 | | Last modified: | 2020-11-25 | | Method: | ELECTRON MICROSCOPY (1.7 Å) | | Cite: | Single-particle cryo-EM at atomic resolution.
Nature, 587, 2020
|
|
1UMD
 
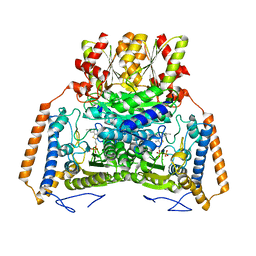 | | branched-chain 2-oxo acid dehydrogenase (E1) from Thermus thermophilus HB8 with 4-methyl-2-oxopentanoate as an intermediate | | Descriptor: | 2-OXO-4-METHYLPENTANOIC ACID, 2-oxo acid dehydrogenase alpha subunit, 2-oxo acid dehydrogenase beta subunit, ... | | Authors: | Nakai, T, Nakagawa, N, Maoka, N, Masui, R, Kuramitsu, S, Kamiya, N, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-09-25 | | Release date: | 2004-03-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Ligand-induced Conformational Changes and a Reaction Intermediate in Branched-chain 2-Oxo Acid Dehydrogenase (E1) from Thermus thermophilus HB8, as Revealed by X-ray Crystallography
J.Mol.Biol., 337, 2004
|
|
1JEH
 
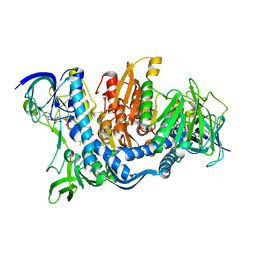 | | CRYSTAL STRUCTURE OF YEAST E3, LIPOAMIDE DEHYDROGENASE | | Descriptor: | DIHYDROLIPOAMIDE DEHYDROGENASE, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Toyoda, T, Suzuki, K, Sekigushi, T, Reed, J, Takenaka, A. | | Deposit date: | 2001-06-18 | | Release date: | 2001-07-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of eucaryotic E3, lipoamide dehydrogenase from yeast.
J.Biochem., 123, 1998
|
|
8JSC
 
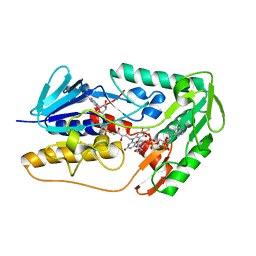 | | Structure of the FSP1 protein from Human | | Descriptor: | 6-HYDROXY-FLAVIN-ADENINE DINUCLEOTIDE, Ferroptosis suppressor protein 1, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Zhang, S.T, Jia, D. | | Deposit date: | 2023-06-19 | | Release date: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | FSP1 oxidizes NADPH to suppress ferroptosis.
Cell Res., 33, 2023
|
|
4M52
 
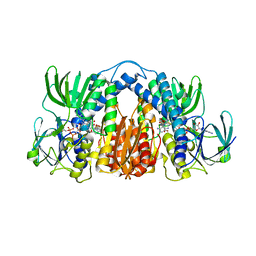 | | Structure of Mtb Lpd bound to SL827 | | Descriptor: | Dihydrolipoyl dehydrogenase, FLAVIN-ADENINE DINUCLEOTIDE, N~2~-[(2-amino-5-bromopyridin-3-yl)sulfonyl]-N-(4-methoxyphenyl)-N~2~-methylglycinamide | | Authors: | Lima, C.D. | | Deposit date: | 2013-08-07 | | Release date: | 2013-11-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Lipoamide channel-binding sulfonamides selectively inhibit mycobacterial lipoamide dehydrogenase.
Biochemistry, 52, 2013
|
|
1RP7
 
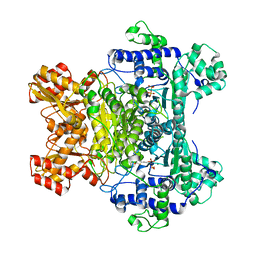 | | E. COLI PYRUVATE DEHYDROGENASE INHIBITOR COMPLEX | | Descriptor: | 2-{3-[(4-AMINO-2-METHYLPYRIMIDIN-5-YL)METHYL]-4-METHYL-2-OXO-2,3-DIHYDRO-1,3-THIAZOL-5-YL}ETHYL TRIHYDROGEN DIPHOSPHATE, MAGNESIUM ION, Pyruvate dehydrogenase E1 component | | Authors: | Arjunan, P, Chandrasekhar, K, Furey, W. | | Deposit date: | 2003-12-03 | | Release date: | 2004-03-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Structural Determinants of Enzyme Binding Affinity: The E1 Component of Pyruvate Dehydrogenase from Escherichia coli in Complex with the Inhibitor Thiamin Thiazolone Diphosphate.
Biochemistry, 43, 2004
|
|
2BFF
 
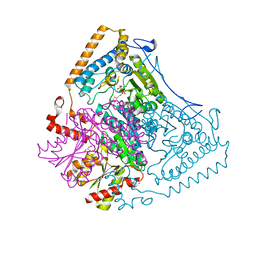 | | Reactivity modulation of human branched-chain alpha-ketoacid dehydrogenase by an internal molecular switch | | Descriptor: | 2-OXOISOVALERATE DEHYDROGENASE ALPHA SUBUNIT, 2-OXOISOVALERATE DEHYDROGENASE BETA SUBUNIT, 2-{3-[(4-AMINO-2-METHYLPYRIMIDIN-5-YL)METHYL]-4-METHYL-2-OXO-2,3-DIHYDRO-1,3-THIAZOL-5-YL}ETHYL TRIHYDROGEN DIPHOSPHATE, ... | | Authors: | Machius, M, Wynn, R.M, Chuang, J.L, Tomchick, D.R, Brautigam, C.A, Chuang, D.T. | | Deposit date: | 2004-12-06 | | Release date: | 2006-02-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | A Versatile Conformational Switch Regulates Reactivity in Human Branched-Chain Alpha-Ketoacid Dehydrogenase.
Structure, 14, 2006
|
|
2BP7
 
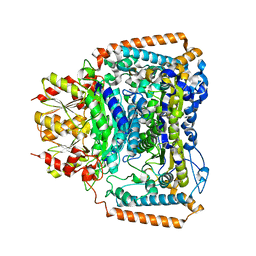 | | New crystal form of the Pseudomonas putida branched-chain dehydrogenase (E1) | | Descriptor: | 2-OXOISOVALERATE DEHYDROGENASE ALPHA SUBUNIT, 2-OXOISOVALERATE DEHYDROGENASE BETA SUBUNIT | | Authors: | Frank, R.A.W, Pratap, J.V, Pei, X.Y, Perham, R.N, Luisi, B.F. | | Deposit date: | 2005-04-18 | | Release date: | 2005-08-10 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The Molecular Origins of Specificity in the Assembly of a Multienzyme Complex.
Structure, 13, 2005
|
|
1V11
 
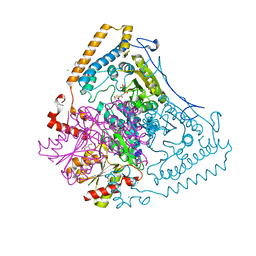 | | CROSSTALK BETWEEN COFACTOR BINDING AND THE PHOSPHORYLATION LOOP CONFORMATION IN THE BCKD MACHINE | | Descriptor: | 2-OXOISOVALERATE DEHYDROGENASE ALPHA SUBUNIT, 2-OXOISOVALERATE DEHYDROGENASE BETA SUBUNIT, BENZAMIDINE, ... | | Authors: | Li, J, Wynn, R.M, Machius, M, Chuang, J.L, Karthikeyan, S, Tomchick, D.R, Chuang, D.T. | | Deposit date: | 2004-04-05 | | Release date: | 2004-06-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Cross-Talk between Thiamin Diphosphate Binding and Phosphorylation Loop Conformation in Human Branched-Chain {Alpha}-Keto Acid Decarboxylase/Dehydrogenase
J.Biol.Chem., 279, 2004
|
|
1UMB
 
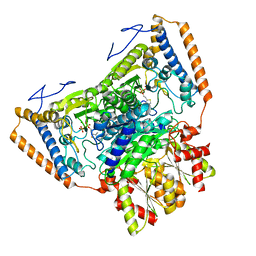 | | branched-chain 2-oxo acid dehydrogenase (E1) from Thermus thermophilus HB8 in holo-form | | Descriptor: | 2-oxo acid dehydrogenase alpha subunit, 2-oxo acid dehydrogenase beta subunit, MAGNESIUM ION, ... | | Authors: | Nakai, T, Nakagawa, N, Maoka, N, Masui, R, Kuramitsu, S, Kamiya, N, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-09-25 | | Release date: | 2004-03-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Ligand-induced Conformational Changes and a Reaction Intermediate in Branched-chain 2-Oxo Acid Dehydrogenase (E1) from Thermus thermophilus HB8, as Revealed by X-ray Crystallography
J.Mol.Biol., 337, 2004
|
|
1V1M
 
 | | CROSSTALK BETWEEN COFACTOR BINDING AND THE PHOSPHORYLATION LOOP CONFORMATION IN THE BCKD MACHINE | | Descriptor: | 2-OXOISOVALERATE DEHYDROGENASE ALPHA SUBUNIT, 2-OXOISOVALERATE DEHYDROGENASE BETA SUBUNIT, BENZAMIDINE, ... | | Authors: | Li, J, Wynn, R.M, Machius, M, Chuang, J.L, Karthikeyan, S, Tomchick, D.R, Chuang, D.T. | | Deposit date: | 2004-04-20 | | Release date: | 2004-06-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Cross-Talk between Thiamin Diphosphate Binding and Phosphorylation Loop Conformation in Human Branched-Chain Alpha-Keto Acid Decarboxylase/Dehydrogenase.
J.Biol.Chem., 279, 2004
|
|
2E75
 
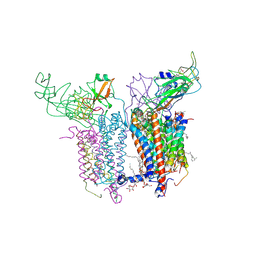 | | Crystal Structure of the Cytochrome b6f Complex with 2-nonyl-4-hydroxyquinoline N-oxide (NQNO) from M.laminosus | | Descriptor: | (7R,17E)-4-HYDROXY-N,N,N,7-TETRAMETHYL-7-[(8E)-OCTADEC-8-ENOYLOXY]-10-OXO-3,5,9-TRIOXA-4-PHOSPHAHEPTACOS-17-EN-1-AMINIUM 4-OXIDE, 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 2-NONYL-4-HYDROXYQUINOLINE N-OXIDE, ... | | Authors: | Cramer, W.A, Yamashita, E, Zhang, H. | | Deposit date: | 2007-01-05 | | Release date: | 2007-06-12 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3.55 Å) | | Cite: | Structure of the Cytochrome b(6)f Complex: Quinone Analogue Inhibitors as Ligands of Heme c(n)
J.Mol.Biol., 370, 2007
|
|
2BEV
 
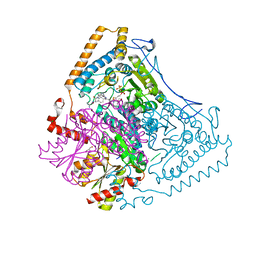 | | Reactivity modulation of human branched-chain alpha-ketoacid dehydrogenase by an internal molecular switch | | Descriptor: | 2-OXOISOVALERATE DEHYDROGENASE ALPHA SUBUNIT, 2-OXOISOVALERATE DEHYDROGENASE BETA SUBUNIT, C2-1-HYDROXY-2-METHYL-BUTYL-THIAMIN DIPHOSPHATE, ... | | Authors: | Machius, M, Wynn, R.M, Chuang, J.L, Tomchick, D.R, Brautigam, C.A, Chuang, D.T. | | Deposit date: | 2004-11-30 | | Release date: | 2006-02-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A Versatile Conformational Switch Regulates Reactivity in Human Branched-Chain Alpha-Ketoacid Dehydrogenase.
Structure, 14, 2006
|
|
2BU6
 
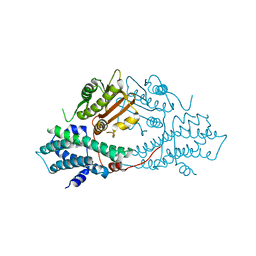 | | crystal structures of human pyruvate dehydrogenase kinase 2 containing physiological and synthetic ligands | | Descriptor: | (N-{4-[(ETHYLANILINO)SULFONYL]-2-METHYLPHENYL}-3,3,3-TRIFLUORO-2-HYDROXY-2-METHYLPROPANAMIDE, PYRUVATE DEHYDROGENASE KINASE ISOENZYME 2 | | Authors: | Knoechel, T.R, Tucker, A.D, Robinson, C.M, Phillips, C, Taylor, W, Bungay, P.J, Kasten, S.A, Roche, T.E, Brown, D.G. | | Deposit date: | 2005-06-08 | | Release date: | 2006-02-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Regulatory Roles of the N-Terminal Domain Based on Crystal Structures of Human Pyruvate Dehydrogenase Kinase 2 Containing Physiological and Synthetic Ligands.
Biochemistry, 45, 2006
|
|
1V1R
 
 | | CROSSTALK BETWEEN COFACTOR BINDING AND THE PHOSPHORYLATION LOOP CONFORMATION IN THE BCKD MACHINE | | Descriptor: | 2-OXOISOVALERATE DEHYDROGENASE ALPHA SUBUNIT, 2-OXOISOVALERATE DEHYDROGENASE BETA SUBUNIT, CHLORIDE ION, ... | | Authors: | Li, J, Wynn, R.M, Machius, M, Chuang, J.L, Karthikeyan, S, Tomchick, D.R, Chuang, D.T. | | Deposit date: | 2004-04-22 | | Release date: | 2004-06-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Cross-Talk between Thiamin Diphosphate Binding and Phosphorylation Loop Conformation in Human Branched-Chain {Alpha}-Keto Acid Decarboxylase/Dehydrogenase
J.Biol.Chem., 279, 2004
|
|
2BU7
 
 | | crystal structures of human pyruvate dehydrogenase kinase 2 containing physiological and synthetic ligands | | Descriptor: | N-(2-AMINOETHYL)-2-{3-CHLORO-4-[(4-ISOPROPYLBENZYL)OXY]PHENYL} ACETAMIDE, PYRUVATE DEHYDROGENASE KINASE ISOENZYME 2 | | Authors: | Knoechel, T.R, Tucker, A.D, Robinson, C.M, Phillips, C, Taylor, W, Bungay, P.J, Kasten, S.A, Roche, T.E, Brown, D.G. | | Deposit date: | 2005-06-08 | | Release date: | 2006-02-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Regulatory Roles of the N-Terminal Domain Based on Crystal Structures of Human Pyruvate Dehydrogenase Kinase 2 Containing Physiological and Synthetic Ligands.
Biochemistry, 45, 2006
|
|
