4A32
 
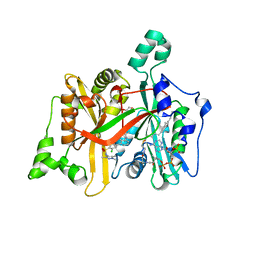 | | CRYSTAL STRUCTURE OF LEISHMANIA MAJOR N-MYRISTOYLTRANSFERASE (NMT) WITH BOUND MYRISTOYL-COA AND A PYRAZOLE SULPHONAMIDE LIGAND | | Descriptor: | 3,5-DICHLORO-3'-[(DIETHYLAMINO)METHYL]-N-(1,3,5-TRIMETHYL-1H-PYRAZOL-4-YL)BIPHENYL-4-SULFONAMIDE, GLYCEROL, GLYCYLPEPTIDE N-TETRADECANOYLTRANSFERASE, ... | | Authors: | Robinson, D.A, Brand, S, Cleghorn, L.A.T, McElroy, S.P, Smith, V.C, Hallyburton, I, Harrison, J.R, Norcross, N.R, Norval, S, Spinks, D, Stojanovski, L, Torrie, L.S, Frearson, J.A, Brenk, R, Fairlamb, A.H, Ferguson, M.A.J, Read, K.D, Wyatt, P.G, Gilbert, I.H. | | Deposit date: | 2011-09-29 | | Release date: | 2011-12-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery of a novel class of orally active trypanocidal N-myristoyltransferase inhibitors.
J. Med. Chem., 55, 2012
|
|
2ZCQ
 
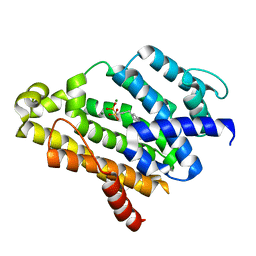 | | Crystal structure of the C(30) carotenoid dehydrosqualene synthase from Staphylococcus aureus complexed with bisphosphonate BPH-652 | | Descriptor: | (1R)-4-(3-phenoxyphenyl)-1-phosphonobutane-1-sulfonic acid, Dehydrosqualene synthase, MAGNESIUM ION | | Authors: | Liu, C.I, Jeng, W.Y, Wang, A.H, Oldfield, E. | | Deposit date: | 2007-11-11 | | Release date: | 2008-03-11 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | A cholesterol biosynthesis inhibitor blocks Staphylococcus aureus virulence.
Science, 319, 2008
|
|
7FUR
 
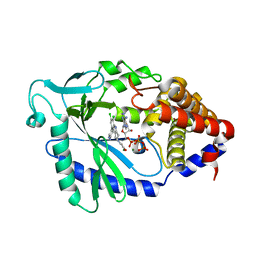 | | Crystal Structure of human cyclic GMP-AMP synthase in complex with 1-[9-(6-aminopyridin-3-yl)-6,7-dichloro-1,3,4,5-tetrahydropyrido[4,3-b]indol-2-yl]-2-hydroxyethanone | | Descriptor: | 1-[9-(6-aminopyridin-3-yl)-6,7-dichloro-1,3,4,5-tetrahydro-2H-pyrido[4,3-b]indol-2-yl]-2-hydroxyethan-1-one, ADENOSINE-5'-TRIPHOSPHATE, Cyclic GMP-AMP synthase, ... | | Authors: | Leibrock, L, Benz, J, Groebke-Zbinden, K, Rudolph, M.G. | | Deposit date: | 2023-02-08 | | Release date: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of a human cyclic GMP-AMP synthase complex
To be published
|
|
4NW4
 
 | |
4CLT
 
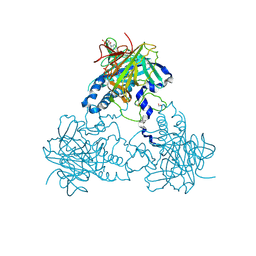 | | Crystal structure of human soluble Adenylyl Cyclase with adenosine-3', 5'-cyclic-monophosphate and pyrophosphate | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, ADENYLATE CYCLASE TYPE 10, ... | | Authors: | Kleinboelting, S, Weyand, M, Steegborn, C. | | Deposit date: | 2014-01-15 | | Release date: | 2014-03-05 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal Structures of Human Soluble Adenylyl Cyclase Reveal Mechanisms of Catalysis and of its Activation Through Bicarbonate.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
5U62
 
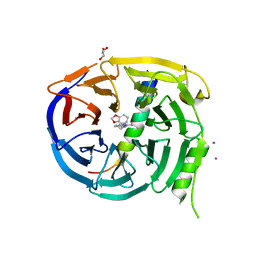 | | Crystal structure of EED in complex with H3K27Me3 peptide and 6-(benzo[d][1,3]dioxol-4-ylmethyl)-5,6,7,8-tetrahydroimidazo[1,5-a]pyridin-3-amine | | Descriptor: | (6S)-6-[(2H-1,3-benzodioxol-4-yl)methyl]-5,6,7,8-tetrahydroimidazo[1,5-a]pyridin-3-amine, GLYCEROL, Histone-lysine N-methyltransferase EZH2, ... | | Authors: | Bussiere, D, Shu, W. | | Deposit date: | 2016-12-07 | | Release date: | 2017-01-11 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-Guided Design of EED Binders Allosterically Inhibiting the Epigenetic Polycomb Repressive Complex 2 (PRC2) Methyltransferase.
J. Med. Chem., 60, 2017
|
|
1HCE
 
 | | STRUCTURE OF HISACTOPHILIN IS SIMILAR TO INTERLEUKIN-1 BETA AND FIBROBLAST GROWTH FACTOR | | Descriptor: | HISACTOPHILIN | | Authors: | Habazettl, J, Gondol, D, Wiltscheck, R, Otlewski, J, Schleicher, M, Holak, T.A. | | Deposit date: | 1994-07-12 | | Release date: | 1994-09-30 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure of hisactophilin is similar to interleukin-1 beta and fibroblast growth factor.
Nature, 359, 1992
|
|
7CM3
 
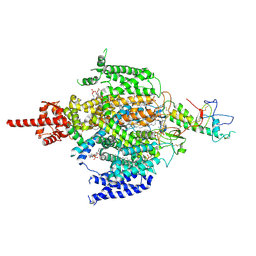 | | Cryo-EM structure of human NALCN in complex with FAM155A | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wu, J, Yan, Z, Ke, M. | | Deposit date: | 2020-07-24 | | Release date: | 2020-11-11 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structure of the human sodium leak channel NALCN in complex with FAM155A.
Nat Commun, 11, 2020
|
|
2QGU
 
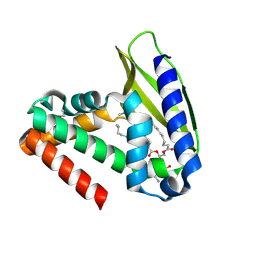 | | Three-dimensional structure of the phospholipid-binding protein from Ralstonia solanacearum Q8XV73_RALSQ in complex with a phospholipid at the resolution 1.53 A. Northeast Structural Genomics Consortium target RsR89 | | Descriptor: | DI-PALMITOYL-3-SN-PHOSPHATIDYLETHANOLAMINE, Probable signal peptide protein | | Authors: | Kuzin, A.P, Chen, Y, Jayaraman, S, Chen, C.X, Fang, Y, Cunningham, K, Ma, L.-C, Xiao, R, Liu, J, Baran, M.C, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-06-29 | | Release date: | 2007-07-24 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Three-dimensional structure of the phospholipid-binding protein from Ralstonia solanacearum Q8XV73_RALSQ in complex with a phospholipid at the resolution 1.53 A.
To be Published
|
|
3O0T
 
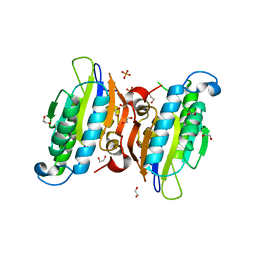 | | Crystal structure of human phosphoglycerate mutase family member 5 (PGAM5) in complex with phosphate | | Descriptor: | 1,2-ETHANEDIOL, PHOSPHATE ION, Serine/threonine-protein phosphatase PGAM5, ... | | Authors: | Chaikuad, A, Alfano, I, Picaud, S, Filippakopoulos, P, Barr, A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Takeda, K, Ichijo, H, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-07-20 | | Release date: | 2010-10-06 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures of PGAM5 Provide Insight into Active Site Plasticity and Multimeric Assembly.
Structure, 25, 2017
|
|
3UTV
 
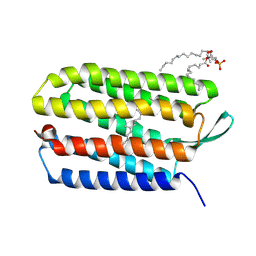 | | Crystal structure of bacteriorhodopsin mutant Y57F | | Descriptor: | 1,2-DIMYRISTOYL-RAC-GLYCERO-3-PHOSPHOCHOLINE, Bacteriorhodopsin, RETINAL | | Authors: | Cao, Z, Bowie, J.U. | | Deposit date: | 2011-11-26 | | Release date: | 2012-05-09 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Shifting hydrogen bonds may produce flexible transmembrane helices.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3Q4P
 
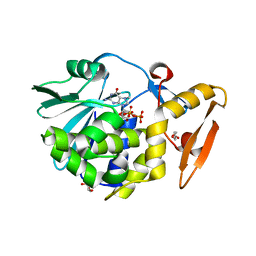 | | Crystal structure of the complex of type I ribosome inactivating protein with 7n-methyl -8-hydroguanosine-5-p-diphosphate at 1.8 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 7N-METHYL-8-HYDROGUANOSINE-5'-DIPHOSPHATE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Kushwaha, G.S, Yamini, S, Sinha, M, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2010-12-24 | | Release date: | 2011-01-12 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the complex of type I ribosome inactivating protein with 7n-methyl-8-hydroguanosine-5-p-diphosphate at 1.8 A resolution
To be Published
|
|
3UO8
 
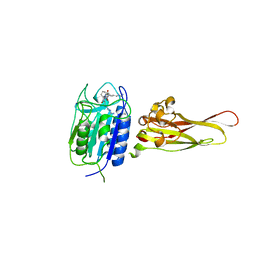 | | Crystal structure of the MALT1 paracaspase (P1 form) | | Descriptor: | Mucosa-associated lymphoid tissue lymphoma translocation protein 1, Z-Val-Arg-Pro-DL-Arg-fluoromethylketone | | Authors: | Jeffrey, P.D, Yu, J.W, Shi, Y. | | Deposit date: | 2011-11-16 | | Release date: | 2011-12-21 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the mucosa-associated lymphoid tissue lymphoma translocation 1 (MALT1) paracaspase region.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3K84
 
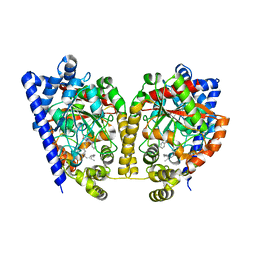 | | Crystal Structure Analysis of a Oleyl/Oxadiazole/pyridine Inhibitor Bound to a Humanized Variant of Fatty Acid Amide Hydrolase | | Descriptor: | (9Z)-1-(5-pyridin-2-yl-1,3,4-oxadiazol-2-yl)octadec-9-en-1-one, CHLORIDE ION, Fatty-acid amide hydrolase 1 | | Authors: | Mileni, M, Stevens, R.C, Boger, D.L. | | Deposit date: | 2009-10-13 | | Release date: | 2009-12-01 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | X-ray crystallographic analysis of alpha-ketoheterocycle inhibitors bound to a humanized variant of fatty acid amide hydrolase.
J.Med.Chem., 53, 2010
|
|
5YU9
 
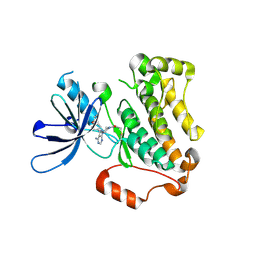 | | Crystal structure of EGFR 696-1022 T790M in complex with Ibrutinib | | Descriptor: | 1-{(3R)-3-[4-amino-3-(4-phenoxyphenyl)-1H-pyrazolo[3,4-d]pyrimidin-1-yl]piperidin-1-yl}prop-2-en-1-one, CHLORIDE ION, Epidermal growth factor receptor | | Authors: | Yan, X.E, Yun, C.H. | | Deposit date: | 2017-11-21 | | Release date: | 2017-12-27 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Ibrutinib targets mutant-EGFR kinase with a distinct binding conformation.
Oncotarget, 7, 2016
|
|
1MDC
 
 | |
4NZ3
 
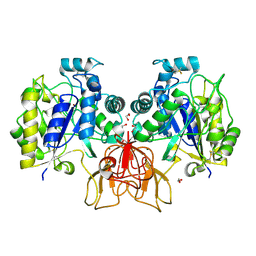 | | Structure of Vibrio cholerae chitin de-N-acetylase in complex with DI(N-ACETYL-D-GLUCOSAMINE) (CBS) in P 21 21 21 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Deacetylase DA1, ... | | Authors: | Albesa-Jove, D, Andres, E, Biarnes, X, Planas, A, Guerin, M.E. | | Deposit date: | 2013-12-11 | | Release date: | 2014-08-13 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.114 Å) | | Cite: | Structural basis of chitin oligosaccharide deacetylation.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
1QMY
 
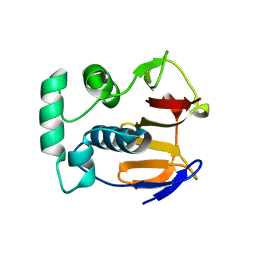 | | FMDV LEADER PROTEASE (LBSHORT-C51A-C133S) | | Descriptor: | 1,2-ETHANEDIOL, PROTEASE | | Authors: | Guarne, A, Tormo, J, Glaser, W, Skern, T, Fita, I. | | Deposit date: | 1999-10-08 | | Release date: | 2000-10-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure and Biochemical Features Distinguish the Foot-and-Mouth Disease Virus Leader Proteinase from Other Papain-Like Enzymes
J.Mol.Biol., 302, 2000
|
|
1IFC
 
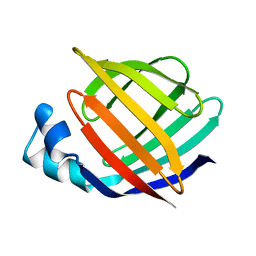 | |
2IF2
 
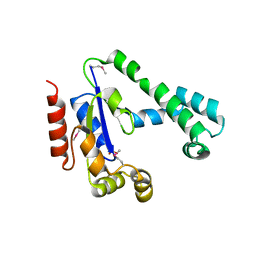 | | Crystal Structure of the Putative Dephospho-CoA Kinase from Aquifex aeolicus, Northeast Structural Genomics Target QR72. | | Descriptor: | 1,2-ETHANEDIOL, Dephospho-CoA kinase, SULFATE ION | | Authors: | Forouhar, F, Hussain, M, Seetharaman, J, Hussain, A, Wu, M, Fang, Y, Cunningham, K, Ma, L.C, Xiao, R, Liu, J, Baran, M, Rost, B, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-09-19 | | Release date: | 2006-10-03 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: |
|
|
3NUQ
 
 | | Structure of a putative nucleotide phosphatase from Saccharomyces cerevisiae | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Dong, A, Yang, C, Singer, A.U, Evdokimova, E, Kudritsdka, M, Brown, G, Edwards, A.M, Joachimiak, A, Savchenko, A, Yakunin, A.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-07-07 | | Release date: | 2010-08-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of a putative nucleotide phosphatase from Saccharomyces cerevisiae
To be Published
|
|
1ZTG
 
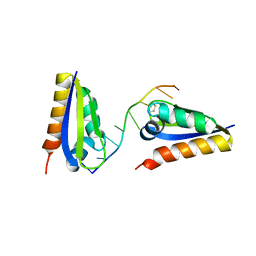 | | human alpha polyC binding protein KH1 | | Descriptor: | 5'-D(P*CP*CP*CP*TP*CP*CP*CP*T)-3', POLY(RC)-BINDING PROTEIN 1 | | Authors: | Sidiqi, M, Wilce, J.A, Barker, A, Schmidgerger, J, Leedman, P.J, Wilce, M.C.J. | | Deposit date: | 2005-05-27 | | Release date: | 2006-05-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Contribution of the first K-homology domain of poly(C)-binding protein 1 to its affinity and specificity for C-rich oligonucleotides
Nucleic Acids Res., 40, 2012
|
|
3O48
 
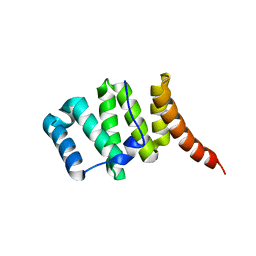 | | Crystal structure of fission protein Fis1 from Saccharomyces cerevisiae | | Descriptor: | Mitochondria fission 1 protein | | Authors: | Tooley, J.E, Khangulov, V, Heroux, A, Bosch, J, Hill, R.B. | | Deposit date: | 2010-07-26 | | Release date: | 2011-08-10 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The 1.75 Angstrom resolution structure of fission protein Fis1 from Saccharomyces cerevisiae reveals elusive interactions of the autoinhibitory domain
Acta Crystallogr.,Sect.F, 67, 2011
|
|
4CGF
 
 | | Interrogating HIV integrase for compounds that bind- a SAMPL challenge | | Descriptor: | 1,2-ETHANEDIOL, 5-{[(3S)-2-oxo-2,3-dihydro-1H-indol-3-yl]methyl}-1,3-benzodioxole-4-carboxylic acid, ACETATE ION, ... | | Authors: | Peat, T.S. | | Deposit date: | 2013-11-23 | | Release date: | 2013-12-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Interrogating HIV Integrase for Compounds that Bind- a Sampl Challenge.
J.Comput.Aided Mol.Des., 28, 2014
|
|
4D0F
 
 | | Human Notch1 EGF domains 11-13 mutant T466A | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, NEUROGENIC LOCUS NOTCH HOMOLOG PROTEIN 1 | | Authors: | Taylor, P, Takeuchi, H, Sheppard, D, Chillakuri, C, Lea, S.M, Haltiwanger, R.S, Handford, P.A. | | Deposit date: | 2014-04-25 | | Release date: | 2014-05-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Fringe-Mediated Extension of O-Linked Fucose in the Ligand-Binding Region of Notch1 Increases Binding to Mammalian Notch Ligands.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
