5O6B
 
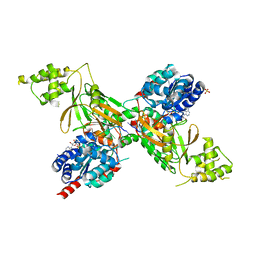 | | Structure of ScPif1 in complex with GGGTTTT and ADP-AlF4 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent DNA helicase PIF1, DNA (5'-D(*GP*GP*GP*TP*TP*T)-3'), ... | | Authors: | Lu, K.Y, Chen, W.F, Rety, S, Liu, N.N, Xu, X.G. | | Deposit date: | 2017-06-06 | | Release date: | 2017-12-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.029 Å) | | Cite: | Insights into the structural and mechanistic basis of multifunctional S. cerevisiae Pif1p helicase.
Nucleic Acids Res., 46, 2018
|
|
7C6H
 
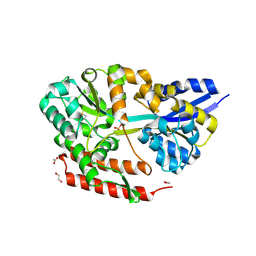 | | Crystal structure of beta-glycosides-binding protein (W177X) of ABC transporter in an open-liganded state bound to laminaribiose | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CHLORIDE ION, ... | | Authors: | Kanaujia, S.P, Chandravanshi, M, Samanta, R. | | Deposit date: | 2020-05-21 | | Release date: | 2020-10-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Conformational Trapping of a beta-Glucosides-Binding Protein Unveils the Selective Two-Step Ligand-Binding Mechanism of ABC Importers.
J.Mol.Biol., 432, 2020
|
|
5O6K
 
 | |
5O9M
 
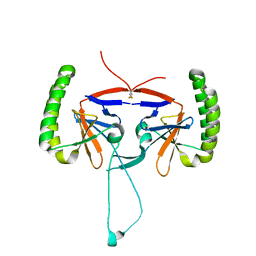 | | Crystal structure of human Histamine-Releasing Factor (HRF/TCTP)containing a disulphide-linked dimer | | Descriptor: | DI(HYDROXYETHYL)ETHER, Translationally-controlled tumor protein | | Authors: | Dore, K.A, Kashiwakura, J, McDonnell, J.M, Gould, H.J, Kawakami, T, Sutton, B.J, Davies, A.M. | | Deposit date: | 2017-06-19 | | Release date: | 2017-12-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structures of murine and human Histamine-Releasing Factor (HRF/TCTP) and a model for HRF dimerisation in mast cell activation.
Mol. Immunol., 93, 2017
|
|
7C6Z
 
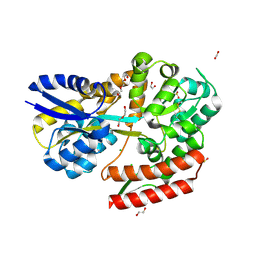 | | Crystal structure of beta-glycosides-binding protein (W67A) of ABC transporter in an open state | | Descriptor: | 1,2-ETHANEDIOL, CARBON DIOXIDE, CHLORIDE ION, ... | | Authors: | Kanaujia, S.P, Chandravanshi, M, Samanta, R. | | Deposit date: | 2020-05-22 | | Release date: | 2020-09-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Conformational Trapping of a beta-Glucosides-Binding Protein Unveils the Selective Two-Step Ligand-Binding Mechanism of ABC Importers.
J.Mol.Biol., 432, 2020
|
|
5O76
 
 | | Structure of phosphoY371 c-CBL in complex with ZAP70-peptide and UbV.pCBL ubiquitin variant | | Descriptor: | CALCIUM ION, E3 ubiquitin-protein ligase CBL, Tyrosine protein kinase ZAP70 peptide, ... | | Authors: | Gabrielsen, M, Buetow, L, Huang, D.T. | | Deposit date: | 2017-06-08 | | Release date: | 2017-11-01 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.473 Å) | | Cite: | A General Strategy for Discovery of Inhibitors and Activators of RING and U-box E3 Ligases with Ubiquitin Variants.
Mol. Cell, 68, 2017
|
|
6S32
 
 | | Crystal structure of ene-reductase CtOYE from Chroococcidiopsis thermalis. | | Descriptor: | ACETATE ION, BENZAMIDINE, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Robescu, M.R, Niero, M, Hall, M, Bergantino, E, Cendron, L. | | Deposit date: | 2019-06-24 | | Release date: | 2020-01-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Two new ene-reductases from photosynthetic extremophiles enlarge the panel of old yellow enzymes: CtOYE and GsOYE.
Appl.Microbiol.Biotechnol., 104, 2020
|
|
5O9V
 
 | | HsNMT1 in complex with CoA and Myristoylated-GGCFSKPK octapeptide | | Descriptor: | Apoptosis-inducing factor 3, CHLORIDE ION, COENZYME A, ... | | Authors: | Dian, C, Meinnel, T, Giglione, C. | | Deposit date: | 2017-06-20 | | Release date: | 2018-06-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.201 Å) | | Cite: | Structural and genomic decoding of human and plant myristoylomes reveals a definitive recognition pattern.
Nat. Chem. Biol., 14, 2018
|
|
7C8B
 
 | | Crystal structure of the SARS-CoV-2 main protease in complex with Z-VAD(OMe)-FMK | | Descriptor: | 3C-like proteinase, CHLORIDE ION, Z-VAD(OMe)-FMK | | Authors: | Zeng, R, Qiao, J.X, Wang, Y.F, Li, Y.S, Yao, R, Yang, S.Y, Lei, J. | | Deposit date: | 2020-05-29 | | Release date: | 2020-09-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the SARS-CoV-2 main protease in complex with Z-VAD(OMe)-FMK
To Be Published
|
|
5OB0
 
 | |
6S73
 
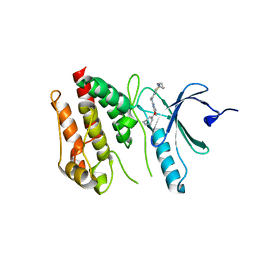 | | Crystal structure of Nek7 SRS mutant bound to compound 51 | | Descriptor: | 3-[[6-(cyclohexylmethoxy)-7~{H}-purin-2-yl]amino]-~{N}-[3-(dimethylamino)propyl]benzenesulfonamide, Serine/threonine-protein kinase Nek7 | | Authors: | Nasir, N, Byrne, M.J, Bhatia, C, Bayliss, R. | | Deposit date: | 2019-07-04 | | Release date: | 2020-06-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Nek7 conformational flexibility and inhibitor binding probed through protein engineering of the R-spine.
Biochem.J., 477, 2020
|
|
5O8Q
 
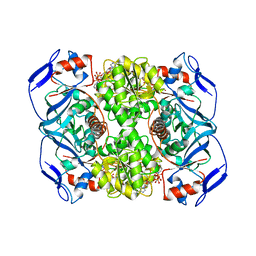 | | Crystal structure of R. ruber ADH-A, mutant Y294F, W295A | | Descriptor: | Alcohol dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ZINC ION | | Authors: | Dobritzsch, D, Maurer, D, Hamnevik, E, Enugala, T.R, Widersten, M. | | Deposit date: | 2017-06-14 | | Release date: | 2017-10-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Relaxation of nonproductive binding and increased rate of coenzyme release in an alcohol dehydrogenase increases turnover with a nonpreferred alcohol enantiomer.
FEBS J., 284, 2017
|
|
7C4I
 
 | |
6S77
 
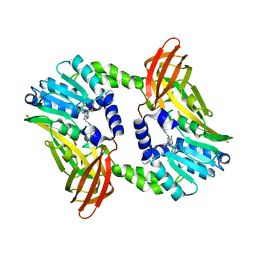 | | Crystal structure of CARM1 N265Y mutant in complex with inhibitor AA183 | | Descriptor: | (2~{S})-4-[[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl-[3-(pyridin-2-ylamino)propyl]amino]-2-azanyl-butanoic acid, Histone-arginine methyltransferase CARM1 | | Authors: | Gunnell, E.A, Muhsen, U, Dowden, J, Dreveny, I. | | Deposit date: | 2019-07-04 | | Release date: | 2020-03-04 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Structural and biochemical evaluation of bisubstrate inhibitors of protein arginine N-methyltransferases PRMT1 and CARM1 (PRMT4).
Biochem.J., 477, 2020
|
|
6RFC
 
 | | Crystal structure of the potassium-pumping G263F mutant of the light-driven sodium pump KR2 in the monomeric form, pH 4.3 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, EICOSANE, GLYCEROL, ... | | Authors: | Kovalev, K, Polovinkin, V, Gushchin, I, Borshchevskiy, V, Gordeliy, V. | | Deposit date: | 2019-04-12 | | Release date: | 2019-04-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and mechanisms of sodium-pumping KR2 rhodopsin.
Sci Adv, 5, 2019
|
|
6S7Y
 
 | |
7C61
 
 | | Crystal structure of 5-HT1B-BRIL and SRP2070_Fab complex | | Descriptor: | 5-hydroxytryptamine receptor 1B,Soluble cytochrome b562,5-hydroxytryptamine receptor 1B, Ergotamine, IGG HEAVY CHAIN, ... | | Authors: | Suzuki, M, Miyagi, H, Asada, H, Yasunaga, M, Suno, C, Takahashi, Y, Saito, J, Iwata, S. | | Deposit date: | 2020-05-21 | | Release date: | 2020-07-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The discovery of a new antibody for BRIL-fused GPCR structure determination.
Sci Rep, 10, 2020
|
|
6RGF
 
 | | Crystal structure of NAD kinase 1 from Listeria monocytogenes in complex with an inhibitor | | Descriptor: | CITRIC ACID, NAD kinase 1, [(2~{R},3~{R},4~{R},5~{R})-2-[8-[3-[[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl-methyl-amino]prop-1-ynyl]-6-azanyl-purin-9-yl]-5-(hydroxymethyl)-4-oxidanyl-oxolan-3-yl] dihydrogen phosphate | | Authors: | Gelin, M, Labesse, G. | | Deposit date: | 2019-04-16 | | Release date: | 2020-05-13 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of NAD kinase 1 from Listeria monocytogenes in complex with an inhibitor
To Be Published
|
|
7C6N
 
 | | Crystal structure of beta-glycosides-binding protein (W177X) of ABC transporter in a closed state bound to cellotetraose (Form II) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, SULFITE ION, ... | | Authors: | Kanaujia, S.P, Chandravanshi, M, Samanta, R. | | Deposit date: | 2020-05-21 | | Release date: | 2020-09-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Conformational Trapping of a beta-Glucosides-Binding Protein Unveils the Selective Two-Step Ligand-Binding Mechanism of ABC Importers.
J.Mol.Biol., 432, 2020
|
|
6S82
 
 | | Structure Of D80A-Fructofuranosidase From Xanthophyllomyces Dendrorhous Complexed With Tris-buffer molecule And hydroquinone | | Descriptor: | 1,2-ETHANEDIOL, 1,4-benzoquinone, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Ramirez-Escudero, M, Sanz-Aparicio, J. | | Deposit date: | 2019-07-08 | | Release date: | 2020-04-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Deciphering the molecular specificity of phenolic compounds as inhibitors or glycosyl acceptors of beta-fructofuranosidase from Xanthophyllomyces dendrorhous.
Sci Rep, 9, 2019
|
|
6RF0
 
 | | Crystal structure of the light-driven sodium pump KR2 in the pentameric "dry" form | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, EICOSANE, RETINAL, ... | | Authors: | Kovalev, K, Polovinkin, V, Gushchin, I, Borshchevskiy, V, Gordeliy, V. | | Deposit date: | 2019-04-12 | | Release date: | 2019-04-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure and mechanisms of sodium-pumping KR2 rhodopsin.
Sci Adv, 5, 2019
|
|
7C8A
 
 | |
7B57
 
 | | Crystal structure of CaMKII-actinin complex bound to ADP | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ADENOSINE-5'-DIPHOSPHATE, Alpha-actinin-2, ... | | Authors: | Zhu, J, Gold, M. | | Deposit date: | 2020-12-03 | | Release date: | 2022-06-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of CaMKII-actinin complex bound to ADP
To Be Published
|
|
5OGF
 
 | | Cu nitrite reductase serial data at varying temperatures RT 0.27MGy | | Descriptor: | ACETATE ION, COPPER (II) ION, Copper-containing nitrite reductase, ... | | Authors: | Horrell, S, Kekilli, D, Strange, R.W, Hough, M.A. | | Deposit date: | 2017-07-13 | | Release date: | 2018-05-23 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Enzyme catalysis captured using multiple structures from one crystal at varying temperatures.
IUCrJ, 5, 2018
|
|
7B7A
 
 | | ENDO-POLYGALACTURONASE FROM ARABIDOPSIS THALIANA | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Pectin lyase-like superfamily protein, alpha-D-mannopyranose-(1-3)-alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Safran, J, Tabi, W, Habrylo, O, Bouckaert, J, Lefebvre, V, Senechal, F, Pelloux, J. | | Deposit date: | 2020-12-10 | | Release date: | 2022-06-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Plant polygalacturonase structures specify enzyme dynamics and processivities to fine-tune cell wall pectins.
Plant Cell, 2023
|
|
