7OSA
 
 | | Pre-translocation complex of 80 S.cerevisiae ribosome with eEF2 and ligands | | Descriptor: | 18S rRNA, 25S rRNA, 40S ribosomal protein S0, ... | | Authors: | Djumagulov, M, Jenner, L, Rozov, A, Demeshkina, N, Yusupov, M, Yusupova, G. | | Deposit date: | 2021-06-08 | | Release date: | 2021-12-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Accuracy mechanism of eukaryotic ribosome translocation.
Nature, 600, 2021
|
|
1MDZ
 
 | | Crystal structure of ArnB aminotransferase with cycloserine and pyridoxal 5' phosphate | | Descriptor: | ArnB aminotransferase, D-[3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YLMETHYL]-N,O-CYCLOSERYLAMIDE, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Noland, B.W, Newman, J.M, Hendle, J, Badger, J, Christopher, J.A, Tresser, J, Buchanan, M.D, Wright, T, Rutter, M.E, Sanderson, W.E, Muller-Dieckmann, H.-J, Gajiwala, K.S, Sauder, J.M, Buchanan, S.G. | | Deposit date: | 2002-08-07 | | Release date: | 2002-12-11 | | Last modified: | 2018-12-26 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Structural studies of Salmonella typhimurium ArnB (PmrH) aminotransferase: A 4-amino-4-deoxy-L-arabinose lipopolysaccharide modifying enzyme
Structure, 10, 2002
|
|
1MHZ
 
 | | METHANE MONOOXYGENASE HYDROXYLASE | | Descriptor: | FE (III) ION, METHANE MONOOXYGENASE HYDROXYLASE | | Authors: | Elango, N, Radhakrishnan, R, Froland, W.A, Waller, B.J, Earhart, C.A, Lipscomb, J.D, Ohlendorf, D.H. | | Deposit date: | 1996-10-21 | | Release date: | 1997-05-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of the hydroxylase component of methane monooxygenase from Methylosinus trichosporium OB3b
Protein Sci., 6, 1997
|
|
4R45
 
 | | Racemic crystal structure of a bimolecular DNA G-quadruplex (P-1) | | Descriptor: | 5'-D(*GP*GP*GP*GP*TP*TP*TP*TP*GP*GP*GP*G)-3', POTASSIUM ION | | Authors: | Mandal, P.K, Collie, G.W, Kauffmann, B, Huc, I. | | Deposit date: | 2014-08-19 | | Release date: | 2014-11-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Racemic DNA crystallography.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
7OSM
 
 | | Intermediate translocation complex of 80 S.cerevisiae ribosome with eEF2 and ligands | | Descriptor: | 18S rRNA, 25S rRNA, 40S ribosomal protein S0, ... | | Authors: | Djumagulov, M, Jenner, L, Rozov, A, Demeshkina, N, Yusupov, M, Yusupova, G. | | Deposit date: | 2021-06-09 | | Release date: | 2021-12-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Accuracy mechanism of eukaryotic ribosome translocation.
Nature, 600, 2021
|
|
4P09
 
 | | Crystal structure of HOIP PUB domain | | Descriptor: | E3 ubiquitin-protein ligase RNF31 | | Authors: | Akutsu, M, Schaeffer, V, Olma, M.H, Gomes, L.C, Kawasaki, M, Dikic, I. | | Deposit date: | 2014-02-20 | | Release date: | 2014-05-07 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Binding of OTULIN to the PUB domain of HOIP controls NF-kappa B signaling.
Mol.Cell, 54, 2014
|
|
4IL8
 
 | | Crystal structure of an H329A mutant of p. aeruginosa PMM/PGM | | Descriptor: | GLYCEROL, MAGNESIUM ION, Phosphomannomutase/phosphoglucomutase | | Authors: | Lee, Y, Mehra-Chaudhary, R, Furdui, C, Beamer, L. | | Deposit date: | 2012-12-29 | | Release date: | 2013-08-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Identification of an essential active-site residue in the alpha-D-phosphohexomutase enzyme superfamily.
Febs J., 280, 2013
|
|
1MOH
 
 | | FERRIC MONOMERIC HEMOGLOBIN I (HB I) | | Descriptor: | HYDROSULFURIC ACID, MONOMERIC HEMOGLOBIN I, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Rizzi, M, Wittenberg, J.B, Ascenzi, P, Bolognesi, M. | | Deposit date: | 1996-02-27 | | Release date: | 1996-08-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural bases for sulfide recognition in Lucina pectinata hemoglobin I.
J.Mol.Biol., 258, 1996
|
|
4IE1
 
 | | Crystal structure of human Arginase-1 complexed with inhibitor 1h | | Descriptor: | Arginase-1, MANGANESE (II) ION, [(5R)-5-amino-5-carboxy-8-hydroxyoctyl](trihydroxy)borate(1-) | | Authors: | Cousido-Siah, A, Mitschler, A, Ruiz, F.X, Beckett, P, Van Zandt, M.C, Ji, M.K, Whitehouse, D, Ryder, T, Jagdmann, E, Andreoli, M, Mazur, A, Padmanilayam, M, Schroeter, H, Golebiowski, A, Podjarny, A. | | Deposit date: | 2012-12-13 | | Release date: | 2013-03-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.0006 Å) | | Cite: | 2-Substituted-2-amino-6-boronohexanoic acids as arginase inhibitors.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
4P0A
 
 | | Crystal structure of HOIP PUB domain in complex with p97 PIM | | Descriptor: | E3 ubiquitin-protein ligase RNF31, Transitional endoplasmic reticulum ATPase | | Authors: | Akutsu, M, Schaeffer, V, Olma, M.H, Gomes, L.C, Kawasaki, M, Dikic, I. | | Deposit date: | 2014-02-20 | | Release date: | 2014-05-07 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3001 Å) | | Cite: | Binding of OTULIN to the PUB domain of HOIP controls NF-kappa B signaling.
Mol.Cell, 54, 2014
|
|
1MHY
 
 | | METHANE MONOOXYGENASE HYDROXYLASE | | Descriptor: | FE (III) ION, METHANE MONOOXYGENASE HYDROXYLASE | | Authors: | Elango, N, Radhakrishnan, R, Froland, W.A, Waller, B.J, Earhart, C.A, Lipscomb, J.D, Ohlendorf, D.H. | | Deposit date: | 1996-10-21 | | Release date: | 1997-05-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the hydroxylase component of methane monooxygenase from Methylosinus trichosporium OB3b
Protein Sci., 6, 1997
|
|
8FMF
 
 | | Structure of CBASS Cap5 from Pseudomonas syringae as an activated tetramer with the cyclic dinucleotide 3'2'-c-diAMP ligand (1 tetramer in the AU) | | Descriptor: | Cyclic (adenosine-(2'-5')-monophosphate-adenosine-(3'-5')-monophosphate, GLYCEROL, HEXAETHYLENE GLYCOL, ... | | Authors: | Rechkoblit, O, Kreitler, D.F, Aggarwal, A.K. | | Deposit date: | 2022-12-23 | | Release date: | 2024-02-07 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Activation of CBASS Cap5 endonuclease immune effector by cyclic nucleotides.
Nat.Struct.Mol.Biol., 31, 2024
|
|
1W2U
 
 | | X-RAY CRYSTAL STRUCTURE OF THE CATALYTIC DOMAIN OF HUMICOLA GRISEA CEL12A IN COMPLEX WITH A SOAKED THIO CELLOTETRAOSE | | Descriptor: | ENDOGLUCANASE, SULFATE ION, TETRAETHYLENE GLYCOL, ... | | Authors: | Berglund, G.I, Shaw, A, Stahlberg, J, Kenne, L, Driguez, T.H, Mitchinson, C, Sandgren, M. | | Deposit date: | 2004-07-08 | | Release date: | 2004-09-16 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Crystal Complex Structures Reveal How Substrate is Bound in the -4 to the +2 Binding Sites of Humicola Grisea Cel12A
J.Mol.Biol., 342, 2004
|
|
2QKX
 
 | | N-acetyl glucosamine 1-phosphate uridyltransferase from Mycobacterium tuberculosis complex with N-acetyl glucosamine 1-phosphate | | Descriptor: | 2-acetamido-2-deoxy-1-O-phosphono-alpha-D-glucopyranose, Bifunctional protein glmU | | Authors: | Zhang, Z, Squire, C.J, Baker, E.N, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2007-07-11 | | Release date: | 2008-07-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structure and function of GlmU from Mycobacterium tuberculosis.
Acta Crystallogr.,Sect.D, 65, 2009
|
|
2QOB
 
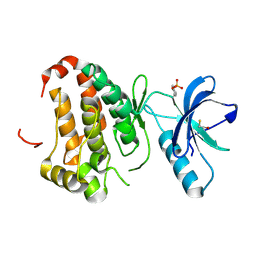 | | Human EphA3 kinase domain, base structure | | Descriptor: | BETA-MERCAPTOETHANOL, Ephrin receptor | | Authors: | Davis, T, Walker, J.R, Newman, E.M, Mackenzie, F, Butler-Cole, C, Weigelt, J, Sundstrom, M, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-07-20 | | Release date: | 2007-08-21 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Autoregulation by the Juxtamembrane Region of the Human Ephrin Receptor Tyrosine Kinase A3 (EphA3).
Structure, 16, 2008
|
|
8FM1
 
 | |
8FMG
 
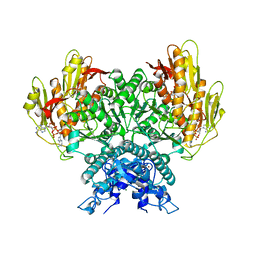 | | Structure of CBASS Cap5 from Pseudomonas syringae as an activated tetramer with the cyclic dinucleotide 3'2'-c-diAMP ligand (3 tetramers in the AU) | | Descriptor: | Cyclic (adenosine-(2'-5')-monophosphate-adenosine-(3'-5')-monophosphate, MAGNESIUM ION, SAVED domain-containing protein, ... | | Authors: | Rechkoblit, O, Kreitler, D.F, Aggarwal, A.K. | | Deposit date: | 2022-12-23 | | Release date: | 2024-02-07 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Activation of CBASS Cap5 endonuclease immune effector by cyclic nucleotides.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8FMH
 
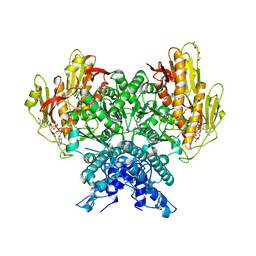 | | Structure of CBASS Cap5 from Pseudomonas syringae as an activated tetramer with the cyclic dinucleotide 3'2'-c-dGAMP ligand (2 tetramers in the AU) | | Descriptor: | 3'2'-cGAMP, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Rechkoblit, O, Kreitler, D.F, Aggarwal, A.K. | | Deposit date: | 2022-12-23 | | Release date: | 2024-02-07 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Activation of CBASS Cap5 endonuclease immune effector by cyclic nucleotides.
Nat.Struct.Mol.Biol., 31, 2024
|
|
4O89
 
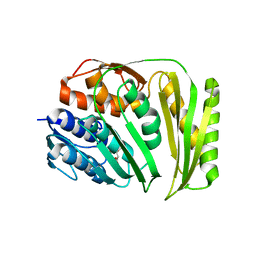 | | Crystal structure of RtcA, the RNA 3'-terminal phosphate cyclase from Pyrococcus horikoshii. | | Descriptor: | CITRIC ACID, RNA 3'-terminal phosphate cyclase | | Authors: | Desai, K.K, Bingman, C.A, Phillips Jr, G.N, Raines, R.T. | | Deposit date: | 2013-12-26 | | Release date: | 2014-09-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of RNA 3'-phosphate cyclase bound to substrate RNA.
Rna, 20, 2014
|
|
6A84
 
 | | Tankyrase-2 in complex with compound 15d | | Descriptor: | 2-(4-chloro-1,2-dihydro-1'H-spiro[indole-3,4'-piperidin]-1'-yl)-5,6,7,8-tetrahydroquinazolin-4(3H)-one, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Niwa, H, Shirai, F, Sato, S, Yoshimoto, N, Tsumura, T, Okue, M, Shirouzu, M, Seimiya, H, Umehara, T. | | Deposit date: | 2018-07-06 | | Release date: | 2019-04-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Discovery of Novel Spiroindoline Derivatives as Selective Tankyrase Inhibitors.
J. Med. Chem., 62, 2019
|
|
1DJW
 
 | | PHOSPHOINOSITIDE-SPECIFIC PHOSPHOLIPASE C-DELTA1 FROM RAT COMPLEXED WITH INOSITOL-2-METHYLENE-1,2-CYCLIC-MONOPHOSPHONATE | | Descriptor: | ACETATE ION, CALCIUM ION, INOSITOL-2-METHYLENE-1,2-CYCLIC-MONOPHOSPHATE, ... | | Authors: | Essen, L.-O, Perisic, O, Williams, R.L. | | Deposit date: | 1996-08-24 | | Release date: | 1997-07-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structural mapping of the catalytic mechanism for a mammalian phosphoinositide-specific phospholipase C.
Biochemistry, 36, 1997
|
|
4IRB
 
 | | Crystal Structure of Vaccinia Virus Uracil DNA Glycosylase Mutant del171-172D4 | | Descriptor: | CHLORIDE ION, GLYCEROL, SULFATE ION, ... | | Authors: | Schormann, N, Zhukovskaya, N, Sartmatova, D, Nuth, M, Ricciardi, R.P, Chattopadhyay, D. | | Deposit date: | 2013-01-14 | | Release date: | 2014-02-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Mutations at the dimer interface affect both function and structure of the Vaccinia virus uracil DNA glycosylase
To be Published
|
|
1VFY
 
 | |
5XI7
 
 | | Crystal structure of T2R-TTL bound with PO-7 | | Descriptor: | (6Z)-3-[[2,5-bis(fluoranyl)phenyl]methylidene]-6-[(4-tert-butyl-1H-imidazol-5-yl)methylidene]piperazine-2,5-dione, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Chu, Y, Wang, Y, Yang, J, Li, W. | | Deposit date: | 2017-04-26 | | Release date: | 2017-10-18 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Synthesis, biological evaluation and X-ray structure of anti-microtubule agents
To Be Published
|
|
5XKF
 
 | | Crystal structure of T2R-TTL-MPC6827 complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, GLYCEROL, ... | | Authors: | Wang, Y, Yang, J, Wang, T, Chen, L. | | Deposit date: | 2017-05-07 | | Release date: | 2018-04-18 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Identification of a powerful and reversible microtubule-inhibitor with efficacy against multidrug-resistant tumors
To Be Published
|
|
