6WXG
 
 | | Cryo-EM reconstruction of VP5*/VP8* assembly from rhesus rotavirus particles - Reversed conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Intermediate capsid protein VP6, ... | | Authors: | Herrmann, T, Harrison, S.C, Jenni, S. | | Deposit date: | 2020-05-10 | | Release date: | 2021-01-20 | | Last modified: | 2021-03-10 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Functional refolding of the penetration protein on a non-enveloped virus.
Nature, 590, 2021
|
|
8AKY
 
 | | 290 A SynPspA rod after incubation with ATP | | Descriptor: | Chloroplast membrane-associated 30 kD protein | | Authors: | Junglas, B, Hudina, E, Schoennenbeck, P, Ritter, I, Santiago-Schuebel, B, Huesgen, P, Sachse, C. | | Deposit date: | 2022-07-31 | | Release date: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Non-canonical ATPase activity drives PspA membrane constriction
To Be Published
|
|
8AKZ
 
 | | 320 A SynPspA rod after incubation with ATP | | Descriptor: | Chloroplast membrane-associated 30 kD protein | | Authors: | Junglas, B, Hudina, E, Schoennenbeck, P, Ritter, I, Santiago-Schuebel, B, Huesgen, P, Sachse, C. | | Deposit date: | 2022-07-31 | | Release date: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (6.9 Å) | | Cite: | Non-canonical ATPase activity drives PspA membrane constriction
To Be Published
|
|
8AKQ
 
 | | 180 A SynPspA rod after incubation with ATP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Chloroplast membrane-associated 30 kD protein | | Authors: | Junglas, B, Hudina, E, Schoennenbeck, P, Ritter, I, Santiago-Schuebel, B, Huesgen, P, Sachse, C. | | Deposit date: | 2022-07-31 | | Release date: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Non-canonical ATPase activity drives PspA membrane constriction
To Be Published
|
|
8AKR
 
 | | 200 A SynPspA rod after incubation with ATP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Chloroplast membrane-associated 30 kD protein | | Authors: | Junglas, B, Hudina, E, Schoennenbeck, P, Ritter, I, Santiago-Schuebel, B, Huesgen, P, Sachse, C. | | Deposit date: | 2022-07-31 | | Release date: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Non-canonical ATPase activity drives PspA membrane constriction
To Be Published
|
|
8AKT
 
 | | 235 A SynPspA rod after incubation with ATP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Chloroplast membrane-associated 30 kD protein | | Authors: | Junglas, B, Hudina, E, Schoennenbeck, P, Ritter, I, Santiago-Schuebel, B, Huesgen, P, Sachse, C. | | Deposit date: | 2022-07-31 | | Release date: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Non-canonical ATPase activity drives PspA membrane constriction
To Be Published
|
|
8AKS
 
 | | 215 A SynPspA rod after incubation with ATP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Chloroplast membrane-associated 30 kD protein | | Authors: | Junglas, B, Hudina, E, Schoennenbeck, P, Ritter, I, Santiago-Schuebel, B, Huesgen, P, Sachse, C. | | Deposit date: | 2022-07-31 | | Release date: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Non-canonical ATPase activity drives PspA membrane constriction
To Be Published
|
|
8AL0
 
 | | 365 A SynPspA rod after incubation with ATP | | Descriptor: | Chloroplast membrane-associated 30 kD protein | | Authors: | Junglas, B, Hudina, E, Schoennenbeck, P, Ritter, I, Santiago-Schuebel, B, Huesgen, P, Sachse, C. | | Deposit date: | 2022-07-31 | | Release date: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (6.9 Å) | | Cite: | Non-canonical ATPase activity drives PspA membrane constriction
To Be Published
|
|
8AKX
 
 | | 305 A SynPspA rod after incubation with ATP | | Descriptor: | Chloroplast membrane-associated 30 kD protein | | Authors: | Junglas, B, Hudina, E, Schoennenbeck, P, Ritter, I, Santiago-Schuebel, B, Huesgen, P, Sachse, C. | | Deposit date: | 2022-07-31 | | Release date: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (6.6 Å) | | Cite: | Non-canonical ATPase activity drives PspA membrane constriction
To Be Published
|
|
8AKV
 
 | | 270 A SynPspA rod after incubation with ATP | | Descriptor: | Chloroplast membrane-associated 30 kD protein | | Authors: | Junglas, B, Hudina, E, Schoennenbeck, P, Ritter, I, Santiago-Schuebel, B, Huesgen, P, Sachse, C. | | Deposit date: | 2022-07-31 | | Release date: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Non-canonical ATPase activity drives PspA membrane constriction
To Be Published
|
|
8AM6
 
 | |
6VYX
 
 | | Escherichia coli transcription-translation complex C4 (TTC-C4) containing mRNA with a 21 nt long spacer, transcription factor NusG, and fMet-tRNAs at P-site and E-site | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S1, ... | | Authors: | Molodtsov, V, Wang, C, Su, M, Ebright, R.H. | | Deposit date: | 2020-02-27 | | Release date: | 2020-09-02 | | Last modified: | 2020-09-23 | | Method: | ELECTRON MICROSCOPY (9.9 Å) | | Cite: | Structural basis of transcription-translation coupling.
Science, 369, 2020
|
|
8AKW
 
 | | 280 A SynPspA rod after incubation with ATP | | Descriptor: | Chloroplast membrane-associated 30 kD protein | | Authors: | Junglas, B, Hudina, E, Schoennenbeck, P, Ritter, I, Santiago-Schuebel, B, Huesgen, P, Sachse, C. | | Deposit date: | 2022-07-31 | | Release date: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (5.4 Å) | | Cite: | Non-canonical ATPase activity drives PspA membrane constriction
To Be Published
|
|
6VZQ
 
 | | Engineered TTLL6 mutant bound to alpha-elongation analog | | Descriptor: | (2~{S})-2-[[[(1~{R})-1-acetamido-4-oxidanyl-4-oxidanylidene-butyl]-phosphonooxy-phosphoryl]methyl]pentanedioic acid, (2~{S})-2-[[[(1~{S})-1-acetamidoethyl]-phosphonooxy-phosphoryl]methyl]pentanedioic acid, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Mahalingan, K.K, Keenen, E.K, Strickland, E.K, Li, Y, Liu, Y, Ball, H.L, Tanner, M.E, Tjandra, N, Roll-Mecak, A. | | Deposit date: | 2020-02-28 | | Release date: | 2020-08-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.08 Å) | | Cite: | Structural basis for polyglutamate chain initiation and elongation by TTLL family enzymes.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6W2C
 
 | | Anomalous iodine signal reveals the position of I-paroxetine complexed with the serotonin transporter at the central site | | Descriptor: | 8B6 heavy chain antibody fragment, 8B6 light chain antibody fragment, I-paroxetine, ... | | Authors: | Coleman, J.A, Navratna, V, Yang, D. | | Deposit date: | 2020-03-05 | | Release date: | 2020-03-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (6.3 Å) | | Cite: | Chemical and structural investigation of the paroxetine-human serotonin transporter complex.
Elife, 9, 2020
|
|
8AAV
 
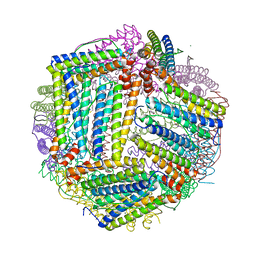 | |
6WAV
 
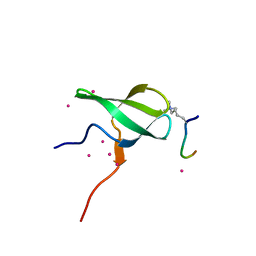 | | Crystal structure of PHF1 in complex with H3K36me3 substitution | | Descriptor: | Histone H3.1, PHD finger protein 1, SULFATE ION, ... | | Authors: | Dong, C, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Min, J.R, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-03-26 | | Release date: | 2020-08-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis for histone variant H3tK27me3 recognition by PHF1 and PHF19.
Elife, 9, 2020
|
|
7TXD
 
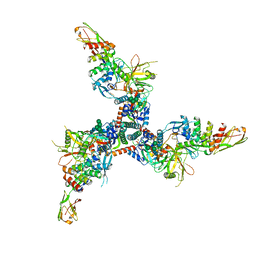 | | Cryo-EM structure of BG505 SOSIP HIV-1 Env trimer in complex with CD4 receptor (D1D2) and broadly neutralizing darpin bnD.9 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Broadly neutralizing darpin bnd.9, ... | | Authors: | Cerutti, G, Gorman, J, Kwong, P.D, Shapiro, L. | | Deposit date: | 2022-02-08 | | Release date: | 2023-04-12 | | Last modified: | 2023-09-27 | | Method: | ELECTRON MICROSCOPY (3.87 Å) | | Cite: | Trapping the HIV-1 V3 loop in a helical conformation enables broad neutralization.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8BON
 
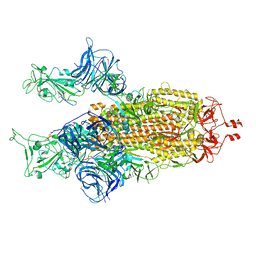 | |
6VWL
 
 | | 70S ribosome bound to HIV frameshifting stem-loop (FSS) and P/E tRNA (rotated conformation) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Loerch, S, Bao, C, Ling, C, Korostelev, A.A, Grigorieff, N, Ermolenko, D.M. | | Deposit date: | 2020-02-20 | | Release date: | 2020-06-03 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | mRNA stem-loops can pause the ribosome by hindering A-site tRNA binding.
Elife, 9, 2020
|
|
6VYQ
 
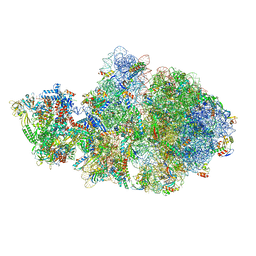 | | Escherichia coli transcription-translation complex A1 (TTC-A1) containing an 15 nt long mRNA spacer, NusG, and fMet-tRNAs at E-site and P-site | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S1, ... | | Authors: | Molodtsov, V, Wang, C, Su, M, Ebright, R.H. | | Deposit date: | 2020-02-27 | | Release date: | 2020-09-02 | | Last modified: | 2020-09-23 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural basis of transcription-translation coupling.
Science, 369, 2020
|
|
6VZ7
 
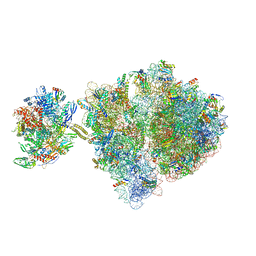 | | Escherichia coli transcription-translation complex C1 (TTC-C1) containing a 27 nt long mRNA spacer, NusG, and fMet-tRNAs at P-site and E-site | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S1, ... | | Authors: | Molodtsov, V, Wang, C, Su, M, Ebright, R.H. | | Deposit date: | 2020-02-27 | | Release date: | 2020-09-02 | | Last modified: | 2020-09-23 | | Method: | ELECTRON MICROSCOPY (7 Å) | | Cite: | Structural basis of transcription-translation coupling.
Science, 369, 2020
|
|
6VZV
 
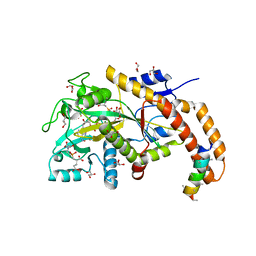 | | TTLL6 bound to gamma-elongation analog | | Descriptor: | (2~{S})-2-[[[(3~{R})-3-acetamido-4-oxidanyl-4-oxidanylidene-butyl]-phosphonooxy-phosphoryl]methyl]pentanedioic acid, (2~{S})-2-[[[(3~{S})-3-acetamido-4-oxidanyl-4-oxidanylidene-butyl]-oxidanyl-phosphoryl]methyl]pentanedioic acid, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Mahalingan, K.K, Keenen, E.K, Strickland, M, Li, Y, Liu, Y, Ball, H.L, Tanner, M.E, Tjandra, N, Roll-Mecak, A. | | Deposit date: | 2020-02-28 | | Release date: | 2020-08-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Structural basis for polyglutamate chain initiation and elongation by TTLL family enzymes.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6W0E
 
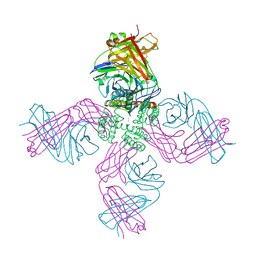 | | Open-gate KcsA soaked in 10 mM BaCl2 | | Descriptor: | BARIUM ION, Fab Heavy Chain, Fab Light Chain, ... | | Authors: | Rohaim, A, Gong, L, Li, J. | | Deposit date: | 2020-02-29 | | Release date: | 2020-07-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.512 Å) | | Cite: | Open and Closed Structures of a Barium-Blocked Potassium Channel.
J.Mol.Biol., 432, 2020
|
|
6W1T
 
 | | RT XFEL structure of Photosystem II 250 microseconds after the second illumination at 2.01 Angstrom resolution | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Ibrahim, M, Fransson, T, Chatterjee, R, Cheah, M.H, Hussein, R, Lassalle, L, Sutherlin, K.D, Young, I.D, Fuller, F.D, Gul, S, Kim, I.-S, Simon, P.S, de Lichtenberg, C, Chernev, P, Bogacz, I, Pham, C, Orville, A.M, Saichek, N, Northen, T.R, Batyuk, A, Carbajo, S, Alonso-Mori, R, Tono, K, Owada, S, Bhowmick, A, Bolotovski, R, Mendez, D, Moriarty, N.W, Holton, J.M, Dobbek, H, Brewster, A.S, Adams, P.D, Sauter, N.K, Bergmann, U, Zouni, A, Messinger, J, Kern, J, Yachandra, V.K, Yano, J. | | Deposit date: | 2020-03-04 | | Release date: | 2020-05-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Untangling the sequence of events during the S2→ S3transition in photosystem II and implications for the water oxidation mechanism.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
