7VWF
 
 | |
7VWE
 
 | |
7VWG
 
 | |
4H4O
 
 | | Crystal Structure of HIV-1 Reverse Transcriptase (RT) in Complex with (E)-3-(3-(2-(2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy)- 4-fluorophenoxy)-5-fluorophenyl)acrylonitrile (JLJ506), A Non-nucleoside inhibitor | | Descriptor: | (2E)-3-(3-{2-[2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy]-4-fluorophenoxy}-5-fluorophenyl)prop-2-enenitrile, Reverse transcriptase/ribonuclease H, Exoribonuclease H, ... | | Authors: | Frey, K.M, Anderson, K.S. | | Deposit date: | 2012-09-17 | | Release date: | 2012-12-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal Structures of HIV-1 Reverse Transcriptase with Picomolar Inhibitors Reveal Key Interactions for Drug Design.
J.Am.Chem.Soc., 134, 2012
|
|
7VWH
 
 | |
2LQ8
 
 | | Domain interaction in Thermotoga maritima NusG | | Descriptor: | Transcription antitermination protein nusG | | Authors: | Droegemueller, J, Stegmann, C, Burmann, B, Roesch, P, Wahl, M.C, Schweimer, K. | | Deposit date: | 2012-02-27 | | Release date: | 2013-01-23 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | An Autoinhibited State in the Structure of Thermotoga maritima NusG.
Structure, 21, 2013
|
|
1HQU
 
 | | HUMAN IMMUNODEFICIENCY VIRUS TYPE 1 | | Descriptor: | (S)-4-ISOPROPOXYCARBONYL-6-METHOXY-3-METHYLTHIOMETHYL-3,4-DIHYDROQUINOXALIN-2(1H)-THIONE, POL POLYPROTEIN | | Authors: | Hsiou, Y, Ding, J, Arnold, E. | | Deposit date: | 2000-12-19 | | Release date: | 2001-05-30 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The Lys103Asn mutation of HIV-1 RT: a novel mechanism of drug resistance.
J.Mol.Biol., 309, 2001
|
|
7VQP
 
 | | Vitamin D receptor complexed with a lithocholic acid derivative | | Descriptor: | 3-((R)-4-((3R,5R,8R,9S,10S,13R,14S,17R)-3-(2-hydroxy-2-methylpropyl)-10,13-dimethylhexadecahydro-1H-cyclopenta[a]phenanthren-17-yl)pentanamido)propanoic acid, Mediator of RNA polymerase II transcription subunit 1, Vitamin D3 receptor | | Authors: | Kato, K, Numoto, N, Kagechika, H, Tanatani, A, Ito, N. | | Deposit date: | 2021-10-20 | | Release date: | 2022-03-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Lithocholic Acid Amides as Potent Vitamin D Receptor Agonists.
Biomolecules, 12, 2022
|
|
7VUE
 
 | | Structural insight of the molecular mechanism of cilofexor bound to FXR | | Descriptor: | 2-[3-[4-[[3-[2,6-bis(chloranyl)phenyl]-5-cyclopropyl-1,2-oxazol-4-yl]methoxy]-2-chloranyl-phenyl]-3-oxidanyl-azetidin-1-yl]pyridine-4-carboxylic acid, Bile acid receptor, Peptide from Nuclear receptor coactivator 2 | | Authors: | Jiang, L, Chen, Y.C. | | Deposit date: | 2021-11-02 | | Release date: | 2022-03-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.601 Å) | | Cite: | Structural insight into the molecular mechanism of cilofexor binding to the farnesoid X receptor.
Biochem.Biophys.Res.Commun., 595, 2022
|
|
2L2O
 
 | |
8R60
 
 | | 1918 H1N1 Viral polymerase heterotrimer in complex with 4 repeat serine-5 phosphorylated PolII peptide | | Descriptor: | Polymerase acidic protein, Polymerase basic protein 2, RNA (5'-R(P*AP*GP*UP*AP*GP*AP*AP*AP*CP*AP*AP*GP*GP*CP*C)-3'), ... | | Authors: | Keown, J.R, Carrique, L, Fodor, E, Grimes, J.M. | | Deposit date: | 2023-11-19 | | Release date: | 2024-03-27 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.23 Å) | | Cite: | Structural and functional characterization of the interaction between the influenza A virus RNA polymerase and the CTD of host RNA polymerase II.
J.Virol., 98, 2024
|
|
4J7N
 
 | | Crystal structure of mouse DXO in complex with M7GPPPG cap | | Descriptor: | 1,2-ETHANEDIOL, 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE-5'-GUANOSINE, 9-METHYLGUANINE, ... | | Authors: | Kilic, T, Chang, J.H, Tong, L. | | Deposit date: | 2013-02-13 | | Release date: | 2013-03-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A mammalian pre-mRNA 5' end capping quality control mechanism and an unexpected link of capping to pre-mRNA processing.
Mol.Cell, 50, 2013
|
|
1EIZ
 
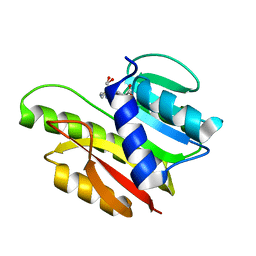 | | FTSJ RNA METHYLTRANSFERASE COMPLEXED WITH S-ADENOSYLMETHIONINE | | Descriptor: | FTSJ, S-ADENOSYLMETHIONINE | | Authors: | Bugl, H, Fauman, E.B, Staker, B.L, Zheng, F, Kushner, S.R, Saper, M.A, Bardwell, J.C.A, Jakob, U. | | Deposit date: | 2000-02-29 | | Release date: | 2000-08-30 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | RNA methylation under heat shock control.
Mol.Cell, 6, 2000
|
|
6TDC
 
 | |
6T4K
 
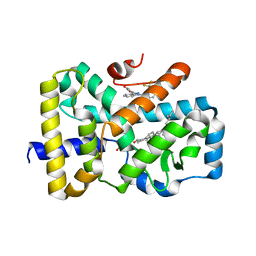 | | ROR(gamma)t ligand binding domain in complex with desmosterol and allosteric ligand MRL871 | | Descriptor: | 4-{1-[2-chloro-6-(trifluoromethyl)benzoyl]-1H-indazol-3-yl}benzoic acid, GLYCEROL, Nuclear receptor ROR-gamma, ... | | Authors: | de Vries, R.M.J.M, Meijer, F.A, Brunsveld, L. | | Deposit date: | 2019-10-14 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Cooperativity between the orthosteric and allosteric ligand binding sites of ROR gamma t.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
6T4W
 
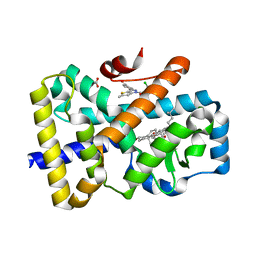 | | ROR(gamma)t ligand binding domain in complex with 20-alpha-hydroxycholesterol and allosteric ligand Glenmark | | Descriptor: | (3alpha,8alpha)-cholest-5-ene-3,20-diol, 4-[1-[2,6-bis(chloranyl)phenyl]carbonyl-5-methyl-thieno[3,2-c]pyrazol-3-yl]benzoic acid, GLYCEROL, ... | | Authors: | de Vries, R.M.J.M, Meijer, F.A, Brunsveld, L. | | Deposit date: | 2019-10-15 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Cooperativity between the orthosteric and allosteric ligand binding sites of ROR gamma t.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
6T4J
 
 | | ROR(gamma)t ligand binding domain in complex with desmosterol and allosteric ligand FM26 | | Descriptor: | 4-[(~{E})-[3-[2-chloranyl-6-(trifluoromethyl)phenyl]-5-(1~{H}-pyrrol-3-yl)-1,2-oxazol-4-yl]methylideneamino]benzoic acid, GLYCEROL, Nuclear receptor ROR-gamma, ... | | Authors: | de Vries, R.M.J.M, Meijer, F.A, Brunsveld, L. | | Deposit date: | 2019-10-14 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Cooperativity between the orthosteric and allosteric ligand binding sites of ROR gamma t.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
6T4X
 
 | | ROR(gamma)t ligand binding domain in complex with 25-hydroxycholesterol and allosteric ligand FM26 | | Descriptor: | 25-HYDROXYCHOLESTEROL, 4-[(~{E})-[3-[2-chloranyl-6-(trifluoromethyl)phenyl]-5-(1~{H}-pyrrol-3-yl)-1,2-oxazol-4-yl]methylideneamino]benzoic acid, GLYCEROL, ... | | Authors: | de Vries, R.M.J.M, Meijer, F.A, Brunsveld, L. | | Deposit date: | 2019-10-15 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Cooperativity between the orthosteric and allosteric ligand binding sites of ROR gamma t.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
4HTT
 
 | |
6T4Y
 
 | | ROR(gamma)t ligand binding domain in complex with 25-hydroxycholesterol and allosteric ligand MRL871 | | Descriptor: | 25-HYDROXYCHOLESTEROL, 4-{1-[2-chloro-6-(trifluoromethyl)benzoyl]-1H-indazol-3-yl}benzoic acid, GLYCEROL, ... | | Authors: | de Vries, R.M.J.M, Meijer, F.A, Brunsveld, L. | | Deposit date: | 2019-10-15 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Cooperativity between the orthosteric and allosteric ligand binding sites of ROR gamma t.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
6TLQ
 
 | | ROR(gamma)t ligand binding domain in complex with cholesterol and allosteric ligand Glenmark | | Descriptor: | 4-[1-[2,6-bis(chloranyl)phenyl]carbonyl-5-methyl-thieno[3,2-c]pyrazol-3-yl]benzoic acid, CHOLESTEROL, GLYCEROL, ... | | Authors: | de Vries, R.M.J.M, Meijer, F.A, Brunsveld, L. | | Deposit date: | 2019-12-03 | | Release date: | 2020-12-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.761 Å) | | Cite: | Cooperativity between the orthosteric and allosteric ligand binding sites of ROR gamma t.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
4I22
 
 | | Structure of the monomeric (V948R)gefitinib/erlotinib resistant double mutant (L858R+T790M) EGFR kinase domain co-crystallized with gefitinib | | Descriptor: | Epidermal growth factor receptor, Gefitinib, SULFATE ION | | Authors: | Gajiwala, K.S, Feng, J, Ferre, R, Ryan, K, Brodsky, O, Stewart, A. | | Deposit date: | 2012-11-21 | | Release date: | 2013-01-16 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Insights into the Aberrant Activity of Mutant EGFR Kinase Domain and Drug Recognition.
Structure, 21, 2013
|
|
6T4G
 
 | | ROR(gamma)t ligand binding domain in complex with cholesterol and allosteric ligand FM26 | | Descriptor: | 4-[(~{E})-[3-[2-chloranyl-6-(trifluoromethyl)phenyl]-5-(1~{H}-pyrrol-3-yl)-1,2-oxazol-4-yl]methylideneamino]benzoic acid, CHOLESTEROL, GLYCEROL, ... | | Authors: | de Vries, R.M.J.M, Meijer, F.A, Brunsveld, L. | | Deposit date: | 2019-10-14 | | Release date: | 2020-11-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Cooperativity between the orthosteric and allosteric ligand binding sites of ROR gamma t.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
1FSI
 
 | | CRYSTAL STRUCTURE OF CYCLIC NUCLEOTIDE PHOSPHODIESTERASE OF APPR>P FROM ARABIDOPSIS THALIANA | | Descriptor: | CYCLIC PHOSPHODIESTERASE, SULFATE ION | | Authors: | Hofmann, A, Zdanov, A, Genschik, P, Filipowicz, W, Ruvinov, S, Wlodawer, A. | | Deposit date: | 2000-09-10 | | Release date: | 2000-11-22 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure and mechanism of activity of the cyclic phosphodiesterase of Appr>p, a product of the tRNA splicing reaction.
EMBO J., 19, 2000
|
|
6GC6
 
 | | 50S ribosomal subunit assembly intermediate state 2 | | Descriptor: | 23S ribosomal RNA, 50S ribosomal protein L13, 50S ribosomal protein L14, ... | | Authors: | Nikolay, R, Hilal, T, Qin, B, Loerke, J, Buerger, J, Mielke, T, Spahn, C.M.T. | | Deposit date: | 2018-04-17 | | Release date: | 2018-07-04 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structural Visualization of the Formation and Activation of the 50S Ribosomal Subunit during In Vitro Reconstitution.
Mol. Cell, 70, 2018
|
|
